Oncofetal TRIM71 drives liver cancer carcinogenesis through remodeling CEBPA-mediated serine/glycine metabolism
- PMID: 39267787
- PMCID: PMC11388079
- DOI: 10.7150/thno.99633
Oncofetal TRIM71 drives liver cancer carcinogenesis through remodeling CEBPA-mediated serine/glycine metabolism
Abstract
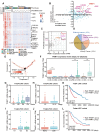
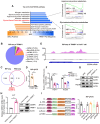
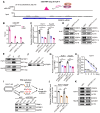
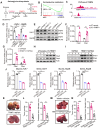
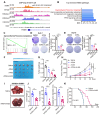
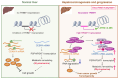
Rationale: Tumor cells remodel transcriptome to construct an ecosystem with stemness features, which maintains tumor growth and highly malignant characteristics. However, the core regulatory factors involved in this process still need to be further discovered. Methods: Single cell RNA-sequncing (scRNA-seq) and bulk RNA-sequencing profiles derived from fetal liver, normal liver, liver tumors, and their adjacent samples were collected to analyze the ecosystem of liver cancer. Mouse models were established to identify molecular functions of oncofetal-related oncogenes using hydrodynamic tail vein injection. Results: We found that liver cancer rebuilt oncofetal ecosystem to maintain malignant features. Interestingly, we identified a group of RNA-binding proteins (RBPs) that were highly overexpressed with oncofetal features. Among them, TRIM71 was specifically expressed in liver cancers and was associated with poor outcomes. TRIM71 drove the carcinogenesis of hepatocellular carcinoma (HCC), and knockdown of TRIM71 significantly abolished liver cancer cell proliferation. Mechanistically, TRIM71 formed a protein complex with IGF2BP1, bound to and stabilized the mRNA of CEBPA in an m6A-dependent manner, enhance the serine/glycine metabolic pathway, and ultimately promoted liver cancer progression. Furthermore, we identified that all-trans-retinoic acid (ATRA) combined with e1A binding protein p300 (EP300) inhibitor A-485 repressed TRIM71, attenuated glycine/serine metabolism, and inhibited liver cancer cell proliferation with high TRIM71 levels. Conclusions: We demonstrated the oncofetal status in liver cancer and highlighted the crucial role of TRIM71 and provided potential therapeutic strategies and liver cancer-specific biomarker for liver cancer patients.
Keywords: CEBPA; RNA-binding proteins; TRIM71; m6A; serine/glycine metabolism.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


Similar articles
-
TRIM71 binds to IMP1 and is capable of positive and negative regulation of target RNAs.Cell Cycle. 2020 Sep;19(18):2314-2326. doi: 10.1080/15384101.2020.1804232. Epub 2020 Aug 20. Cell Cycle. 2020. PMID: 32816599 Free PMC article.
-
Circ_0007429/miR-637/TRIM71/Ago2 axis participates in the regulation of proliferation, migration, invasion, apoptosis, and aerobic glycolysis of HCC.Mol Carcinog. 2023 Jun;62(6):820-832. doi: 10.1002/mc.23526. Epub 2023 Mar 15. Mol Carcinog. 2023. PMID: 36920046
-
Tripartite Motif Containing 71 Suppresses Tumor Growth by Down-Regulating eIF5A2 Expression in Laryngeal Squamous Cell Carcinoma.Appl Biochem Biotechnol. 2025 Mar;197(3):1504-1515. doi: 10.1007/s12010-024-05084-1. Epub 2024 Nov 23. Appl Biochem Biotechnol. 2025. PMID: 39579322
-
Oncofetal SNRPE promotes HCC tumorigenesis by regulating the FGFR4 expression through alternative splicing.Br J Cancer. 2024 Jul;131(1):77-89. doi: 10.1038/s41416-024-02689-5. Epub 2024 May 25. Br J Cancer. 2024. PMID: 38796598 Free PMC article.
-
FOXM1-activated IGF2BP3 promotes cell malignant phenotypes and M2 macrophage polarization in hepatocellular carcinoma by inhibiting ferroptosis via stabilizing RRM2 mRNA in an m6A-dependent manner.Mol Cell Biochem. 2025 May;480(5):3051-3066. doi: 10.1007/s11010-024-05170-2. Epub 2024 Dec 4. Mol Cell Biochem. 2025. PMID: 39630361
Cited by
-
Lyophilized apoptotic vesicles improve hemostasis and bone regeneration in traumatic patients with impacted third molar extraction.Mol Ther. 2025 Jun 4;33(6):2931-2944. doi: 10.1016/j.ymthe.2025.02.033. Epub 2025 Feb 22. Mol Ther. 2025. PMID: 39988872 Clinical Trial.
-
A review of C/EBP α: a potential novel target for solid tumor intervention.J Transl Med. 2025 Aug 11;23(1):894. doi: 10.1186/s12967-025-06884-7. J Transl Med. 2025. PMID: 40790495 Free PMC article. Review.
References
-
- Hanahan D. Hallmarks of cancer: New dimensions. Cancer Discov. 2022;12:31–46. - PubMed
-
- Sharma A, Seow JJW, Dutertre CA, Pai R, Bleriot C, Mishra A. et al. Onco-fetal reprogramming of endothelial cells drives immunosuppressive macrophages in hepatocellular carcinoma. Cell. 2020;183:377–94. e21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

