Stable Platform for Mevalonate Bioproduction from CO2
- PMID: 39268049
- PMCID: PMC11388446
- DOI: 10.1021/acssuschemeng.4c03561
Stable Platform for Mevalonate Bioproduction from CO2
Abstract
Stable production of value-added products using a microbial chassis is pivotal for determining the industrial suitability of the engineered biocatalyst. Microbial cells often lose the multicopy expression plasmids during long-term cultivations. Owing to the advantages related to titers, yields, and productivities when using a multicopy expression system compared with genomic integrations, plasmid stability is essential for industrially relevant biobased processes. Cupriavidus necator H16, a facultative chemolithoautotrophic bacterium, has been successfully engineered to convert inorganic carbon obtained from CO2 fixation into value-added products. The application of this unique capability in the biotech industry has been hindered by C. necator H16 inability to stably maintain multicopy plasmids. In this study, we designed and tested plasmid addiction systems based on the complementation of essential genes. Among these, implementation of a plasmid addiction tool based on the complementation of mutants lacking RubisCO, which is essential for CO2 fixation, successfully stabilized a multicopy plasmid. Expressing the mevalonate pathway operon (MvaES) using this addiction system resulted in the production of ∼10 g/L mevalonate with carbon yields of ∼25%. The mevalonate titers and yields obtained here using CO2 are the highest achieved to date for the production of C6 compounds from C1 feedstocks.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
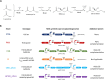


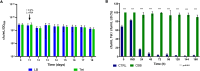
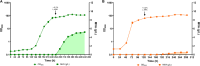

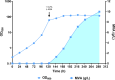
Similar articles
-
Carbon dioxide valorization into resveratrol via lithoautotrophic fermentation using engineered Cupriavidus necator H16.Microb Cell Fact. 2024 Apr 27;23(1):122. doi: 10.1186/s12934-024-02398-x. Microb Cell Fact. 2024. PMID: 38678199 Free PMC article.
-
CO2-based production of phytase from highly stable expression plasmids in Cupriavidus necator H16.Microb Cell Fact. 2024 Jan 3;23(1):9. doi: 10.1186/s12934-023-02280-2. Microb Cell Fact. 2024. PMID: 38172920 Free PMC article.
-
Development of a plasmid-based, tunable, tolC-derived expression system for application in Cupriavidus necator H16.J Biotechnol. 2018 May 20;274:15-27. doi: 10.1016/j.jbiotec.2018.03.007. Epub 2018 Mar 13. J Biotechnol. 2018. PMID: 29549002
-
Problems and corresponding strategies for converting CO2 into value-added products in Cupriavidus necator H16 cell factories.Biotechnol Adv. 2023 Oct;67:108183. doi: 10.1016/j.biotechadv.2023.108183. Epub 2023 Jun 5. Biotechnol Adv. 2023. PMID: 37286176 Review.
-
Design of inducible expression vectors for improved protein production in Ralstonia eutropha H16 derived host strains.J Biotechnol. 2016 Oct 10;235:92-9. doi: 10.1016/j.jbiotec.2016.04.026. Epub 2016 Apr 13. J Biotechnol. 2016. PMID: 27085887 Review.
Cited by
-
Unlocking the potential of Cupriavidus necator H16 as a platform for bioproducts production from carbon dioxide.World J Microbiol Biotechnol. 2024 Nov 22;40(12):389. doi: 10.1007/s11274-024-04200-x. World J Microbiol Biotechnol. 2024. PMID: 39572451 Review.
-
Synthetic Genetic Elements Enable Rapid Characterization of Inorganic Carbon Uptake Systems in Cupriavidus necator H16.ACS Synth Biol. 2025 Mar 21;14(3):943-953. doi: 10.1021/acssynbio.4c00869. Epub 2025 Mar 6. ACS Synth Biol. 2025. PMID: 40048245
-
Efficient Production of High-Concentration Poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) from CO2 Employing the Recombinant of Cupriavidus necator.Bioengineering (Basel). 2025 May 22;12(6):557. doi: 10.3390/bioengineering12060557. Bioengineering (Basel). 2025. PMID: 40564374 Free PMC article.
-
CnRed: Efficient, Marker-free Genome Engineering of Cupriavidus necator H16 by Adapted Lambda Red Recombineering.ACS Synth Biol. 2025 Mar 21;14(3):842-854. doi: 10.1021/acssynbio.4c00757. Epub 2025 Feb 24. ACS Synth Biol. 2025. PMID: 39989320 Free PMC article.
References
-
- Parry M. L.; Rosenzweig C.; Iglesias A.; Livermore M.; Fischer G. Effects of climate change on global food production under SRES emissions and socio-economic scenarios. Global Environ. Change 2004, 14 (1), 53–67. 10.1016/j.gloenvcha.2003.10.008. - DOI
-
- Rosenzweig C.; Iglesias A.; Yang X. B.; Epstein P. R.; Chivian E. Climate Change and Extreme Weather Events; Implications for Food Production, Plant Diseases, and Pests. Global Change Hum. Health 2001, 2 (2), 90–104. 10.1023/A:1015086831467. - DOI
-
- Liew F. E.; Nogle R.; Abdalla T.; Rasor B. J.; Canter C.; Jensen R. O.; Wang L.; Strutz J.; Chirania P.; De Tissera S.; et al. Carbon-negative production of acetone and isopropanol by gas fermentation at industrial pilot scale. Nat. Biotechnol. 2022, 40 (3), 335–344. 10.1038/s41587-021-01195-w. - DOI - PubMed
LinkOut - more resources
Full Text Sources
