Taxonomic composition and functional potentials of gastrointestinal microbiota in 12 wild-stranded cetaceans
- PMID: 39268538
- PMCID: PMC11390675
- DOI: 10.3389/fmicb.2024.1394745
Taxonomic composition and functional potentials of gastrointestinal microbiota in 12 wild-stranded cetaceans
Abstract
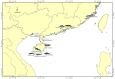
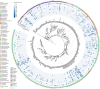
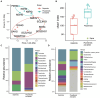
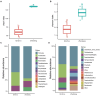
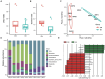
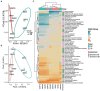
Cetaceans play a crucial role in marine ecosystems; however, research on their gastrointestinal microbiota remains limited due to sampling constraints. In this study, we collected hindgut samples from 12 stranded cetaceans and performed 16S rRNA gene amplicon sequencing to investigate microbial composition and functional potentials. Analysis of ZOTUs profiles revealed that the phyla Firmicutes, Proteobacteria, and Bacteroidetes dominated all hindgut samples. However, unique microbial profiles were observed among different cetacean species, with significant separation of gut microbiota communities according to biological evolutionary lineages. Different genera that contain pathogens were observed distinguishing delphinids from physeteroids/ziphiids. Delphinid samples exhibited higher abundances of Vibrio, Escherichia, and Paeniclostridium, whereas physeteroid and ziphiid samples showed higher abundances of Pseudomonas, Enterococcus, and Intestinimonas. Functional analysis indicated convergence in the gut microbiota among all cetaceans, with shared bacterial infection pathways across hindgut samples. In addition, a comparison of the gastrointestinal microbial composition between a stranded short-finned pilot whale (Globicephala macrorhynchus) and a stranded rough-toothed dolphin (Steno bredanensis) using 16S rRNA gene sequencing revealed distinct microbial community structures and functional capacities. To the best of our knowledge, this study represents the first report on the gastrointestinal microbiota of the pantropical spotted dolphin (Stenella attenuata), Blainville's beaked whale (Mesoplodon densirostris), and rough-toothed dolphin, with various comparisons conducted among different cetacean species. Our findings enhance the understanding of microbial composition and diversity in cetacean gastrointestinal microbiota, providing new insights into co-evolution and complex interactions between cetacean microbes and hosts.
Keywords: delphinids; food digestion; functional potentials; gastrointestinal microbiota; gut microbiota; physeteroids and ziphiid; stranded cetaceans.
Copyright © 2024 Fan, Kang, Lv, Zhai, Jia, Yang, Shi, Zhou, Diao, Li, Jin, Liu, Kristiansen, Zhang, Chen and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Barros N. B., Jefferson T. A., Parsons E. (2004). Feeding habits of indo-Pacific humpback dolphins (Sousa chinensis) stranded in Hong Kong. Aquat. Mamm. 30, 179–188. doi: 10.1578/AM.30.1.2004.179 - DOI
LinkOut - more resources
Full Text Sources

