Salmonella Typhimurium infection inhibits macrophage IFNβ signaling in a TLR4-dependent manner
- PMID: 39269166
- PMCID: PMC11475681
- DOI: 10.1128/iai.00098-24
Salmonella Typhimurium infection inhibits macrophage IFNβ signaling in a TLR4-dependent manner
Abstract
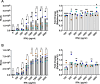
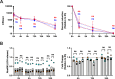
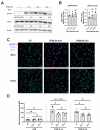
Type I Interferons (IFNs) generally have a protective role during viral infections, but their function during bacterial infections is dependent on the bacterial species. Legionella pneumophila, Shigella sonnei and Mycobacterium tuberculosis can inhibit type I IFN signaling. Here we examined the role of type I IFN, specifically IFNβ, in the context of Salmonella enterica serovar Typhimurium (STm) macrophage infections and the capacity of STm to inhibit type I IFN signaling. We demonstrate that IFNβ has no effect on the intracellular growth of STm in infected bone marrow derived macrophages (BMDMs) derived from C57BL/6 mice. STm infection inhibits IFNβ signaling but not IFNγ signaling in a murine macrophage cell line. We show that this inhibition is independent of the type III and type VI secretion systems expressed by STm and is also independent of bacterial phagocytosis. The inhibition is Toll-like receptor 4 (TLR4)-dependent as the TLR4 ligand, lipopolysaccharide (LPS), alone is sufficient to inhibit IFNβ-mediated signaling. Cells downregulated their surface levels of IFNα/β receptor 1 (IFNAR1) in response to LPS, which may be mediating our observed inhibition. Lastly, we examined this inhibition in the context of TLR4-deficient BMDMs as well as TLR4 RNA interference and we observed a loss of inhibition with LPS stimulation as well as STm infection. In summary, we show that macrophages exposed to STm have reduced IFNβ signaling via crosstalk with TLR4 signaling, which may be mediated by reduced host cell surface IFNAR1, and that IFNβ signaling does not affect cell-autonomous host defense against STm.
Keywords: Salmonella; interferons; macrophages; toll receptors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Update of
-
Salmonella Typhimurium infection inhibits macrophage IFNβ signaling in a TLR4-dependent manner.bioRxiv [Preprint]. 2024 Mar 5:2024.03.05.583530. doi: 10.1101/2024.03.05.583530. bioRxiv. 2024. Update in: Infect Immun. 2024 Oct 15;92(10):e0009824. doi: 10.1128/iai.00098-24. PMID: 38496427 Free PMC article. Updated. Preprint.
References
-
- Hapfelmeier S, Stecher B, Barthel M, Kremer M, Müller AJ, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, Hardt W-D. 2005. The Salmonella pathogenicity island (SPI)-2 and SPI-1 Type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms1. J Immunol 174:1675–1685. doi:10.4049/jimmunol.174.3.1675 - DOI - PubMed
-
- Raffatellu M, Wilson RP, Chessa D, Andrews-Polymenis H, Tran QT, Lawhon S, Khare S, Adams LG, Bäumler AJ. 2005. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype typhimurium invasion of epithelial cells. Infect Immun 73:146–154. doi:10.1128/IAI.73.1.146-154.2005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

