A novel luciferase-based reporter tool to monitor the dynamics of carbon catabolite repression in filamentous fungi
- PMID: 39269439
- PMCID: PMC11395683
- DOI: 10.1111/1751-7915.70012
A novel luciferase-based reporter tool to monitor the dynamics of carbon catabolite repression in filamentous fungi
Abstract
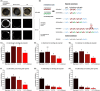
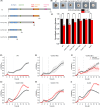
Filamentous fungi with their diverse inventory of carbohydrate-active enzymes promise a holistic usage of lignocellulosic residues. A major challenge for application is the inherent repression of enzyme production by carbon catabolite repression (CCR). In the presence of preferred carbon sources, the transcription factor CreA/CRE-1 binds to specific but conserved motifs in promoters of genes involved in sugar metabolism, but the status of CCR is notoriously difficult to quantify. To allow for a real-time evaluation of CreA/CRE-1-mediated CCR at the transcriptional level, we developed a luciferase-based construct, representing a dynamic, highly responsive reporter system that is inhibited by monosaccharides in a quantitative fashion. Using this tool, CreA/CRE-1-dependent CCR triggered by several monosaccharides could be measured in Neurospora crassa, Aspergillus niger and Aspergillus nidulans over the course of hours, demonstrating distinct and dynamic regulatory processes. Furthermore, we used the reporter to visualize the direct impacts of multiple CreA truncations on CCR induction. Our reporter thus offers a widely applicable quantitative approach to evaluate CreA/CRE-1-mediated CCR across diverse fungal species and will help to elucidate the multifaceted effects of CCR on fungal physiology for both basic research and industrial strain engineering endeavours.
© 2024 The Author(s). Microbial Biotechnology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no competing interests to declare that are relevant to the content of this article.
Figures



Similar articles
-
Carbon Catabolite Repression in Filamentous Fungi Is Regulated by Phosphorylation of the Transcription Factor CreA.mBio. 2021 Jan 5;12(1):e03146-20. doi: 10.1128/mBio.03146-20. mBio. 2021. PMID: 33402538 Free PMC article.
-
Carbon Catabolite Repression Governs Diverse Physiological Processes and Development in Aspergillus nidulans.mBio. 2021 Feb 22;13(1):e0373421. doi: 10.1128/mbio.03734-21. Epub 2022 Feb 15. mBio. 2021. PMID: 35164551 Free PMC article.
-
Regulation of Aspergillus nidulans CreA-Mediated Catabolite Repression by the F-Box Proteins Fbx23 and Fbx47.mBio. 2018 Jun 19;9(3):e00840-18. doi: 10.1128/mBio.00840-18. mBio. 2018. PMID: 29921666 Free PMC article.
-
Implications of carbon catabolite repression for Aspergillus-based cell factories: A review.Biotechnol J. 2024 Feb;19(2):e2300551. doi: 10.1002/biot.202300551. Biotechnol J. 2024. PMID: 38403447 Review.
-
Carbon Catabolite Repression in Filamentous Fungi.Int J Mol Sci. 2017 Dec 24;19(1):48. doi: 10.3390/ijms19010048. Int J Mol Sci. 2017. PMID: 29295552 Free PMC article. Review.
Cited by
-
Engineering of Aspergillus niger for efficient production of D-xylitol from L-arabinose.Microb Cell Fact. 2024 Oct 5;23(1):262. doi: 10.1186/s12934-024-02526-7. Microb Cell Fact. 2024. PMID: 39367393 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

