Endogenous retroviral ERVH48-1 promotes human urine cell reprogramming
- PMID: 39269631
- PMCID: PMC11399365
- DOI: 10.1186/s13619-024-00200-2
Endogenous retroviral ERVH48-1 promotes human urine cell reprogramming
Erratum in
-
Correction: Endogenous retroviral ERVH48-1 promotes human urine cell reprogramming.Cell Regen. 2026 Jan 19;15(1):3. doi: 10.1186/s13619-026-00281-1. Cell Regen. 2026. PMID: 41553632 Free PMC article. No abstract available.
Abstract
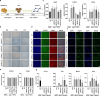
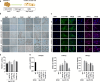
Endogenous retroviruses (ERVs), once thought to be mere remnants of ancient viral integrations in the mammalian genome, are now recognized for their critical roles in various physiological processes, including embryonic development, innate immunity, and tumorigenesis. Their impact on host organisms is significant driver of evolutionary changes, offering insight into evolutionary mechanisms. In our study, we explored the functionality of ERVs by examining single-cell transcriptomic profiles from human embryonic stem cells and urine cells. This led to the discovery of a unique ERVH48-1 expression pattern between these cell types. Additionally, somatic cell reprogramming efficacy was enhanced when ERVH48-1 was overexpressed in a urine cell-reprogramming system. Induced pluripotent stem cells (iPSCs) generated with ERVH48-1 overexpression recapitulated the traits of those produced by traditional reprogramming approaches, and the resulting iPSCs demonstrated the capability to differentiate into all three germ layers in vitro. Our research elucidated the role of ERVs in somatic cell reprogramming.
Keywords: ERVH48-1; Endogenous retroviruses; Human embryonic stem cells; Induced pluripotent stem cells; Urine cell integration-free reprogramming system.
© 2024. The Author(s).
Conflict of interest statement
Duanqing Pei is a member of the Editorial Board for Cell Regeneration. He was not involved in the journal’s review of, or decisions related to, this manuscript.
Figures




References
LinkOut - more resources
Full Text Sources
