A pleiotropic recurrent dominant ITPR3 variant causes a complex multisystemic disease
- PMID: 39270020
- PMCID: PMC11397499
- DOI: 10.1126/sciadv.ado5545
A pleiotropic recurrent dominant ITPR3 variant causes a complex multisystemic disease
Abstract
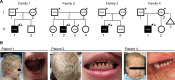
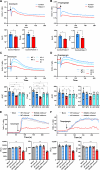
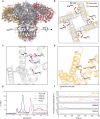
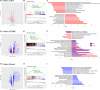
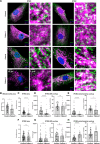
Inositol 1,4,5-trisphosphate (IP3) receptor type 1 (ITPR1), 2 (ITPR2), and 3 (ITPR3) encode the IP3 receptor (IP3R), a key player in intracellular calcium release. In four unrelated patients, we report that an identical ITPR3 de novo variant-NM_002224.3:c.7570C>T, p.Arg2524Cys-causes, through a dominant-negative effect, a complex multisystemic disorder with immunodeficiency. This leads to defective calcium homeostasis, mitochondrial malfunction, CD4+ lymphopenia, a quasi-absence of naïve CD4+ and CD8+ cells, an increase in memory cells, and a distinct TCR repertoire. The calcium defect was recapitulated in Jurkat knock-in. Site-directed mutagenesis displayed the exquisite sensitivity of Arg2524 to any amino acid change. Despite the fact that all patients had severe immunodeficiency, they also displayed variable multisystemic involvements, including ectodermal dysplasia, Charcot-Marie-Tooth disease, short stature, and bone marrow failure. In conclusion, unlike previously reported ITPR1-3 deficiencies leading to narrow, mainly neurological phenotypes, a recurrent dominant ITPR3 variant leads to a multisystemic disease, defining a unique role for IP3R3 in the tetrameric IP3R complex.
Figures


References
-
- Luan S., Wang C., Calcium signaling mechanisms across kingdoms. Annu. Rev. Cell Dev. Biol. 37, 311–340 (2021). - PubMed
-
- Bakowski D., Murray F., Parekh A. B., Store-operated Ca2+ channels: Mechanism, function, pharmacology, and therapeutic targets. Annu. Rev. Pharmacol. Toxicol. 61, 629–654 (2021). - PubMed
-
- Berridge M. J., The inositol trisphosphate/calcium signaling pathway in health and disease. Physiol. Rev. 96, 1261–1296 (2016). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

