Blocking tumor-intrinsic MNK1 kinase restricts metabolic adaptation and diminishes liver metastasis
- PMID: 39270021
- PMCID: PMC11397505
- DOI: 10.1126/sciadv.adi7673
Blocking tumor-intrinsic MNK1 kinase restricts metabolic adaptation and diminishes liver metastasis
Abstract
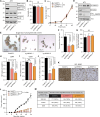
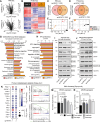
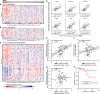
Dysregulation of the mitogen-activated protein kinase interacting kinases 1/2 (MNK1/2)-eukaryotic initiation factor 4E (eIF4E) signaling axis promotes breast cancer progression. MNK1 is known to influence cancer stem cells (CSCs); self-renewing populations that support metastasis, recurrence, and chemotherapeutic resistance, making them a clinically relevant target. The precise function of MNK1 in regulating CSCs, however, remains unexplored. Here, we generated MNK1 knockout cancer cell lines, resulting in diminished CSC properties in vitro and slowed tumor growth in vivo. Using a multiomics approach, we functionally demonstrated that loss of MNK1 restricts tumor cell metabolic adaptation by reducing glycolysis and increasing dependence on oxidative phosphorylation. Furthermore, MNK1-null breast and pancreatic tumor cells demonstrated suppressed metastasis to the liver, but not the lung. Analysis of The Cancer Genome Atlas (TCGA) data from breast cancer patients validated the positive correlation between MNK1 and glycolytic enzyme protein expression. This study defines metabolic perturbations as a previously unknown consequence of targeting MNK1/2, which may be therapeutically exploited.
Figures



References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). - PubMed
-
- American Cancer Society, Cancer Facts & Figures 2020 (American Cancer Society, 2020).
-
- Chu J., Cargnello M., Topisirovic I., Pelletier J., Translation initiation factors: Reprogramming protein synthesis in cancer. Trends Cell Biol. 26, 918–933 (2016). - PubMed
-
- Lazaris-Karatzas A., Montine K. S., Sonenberg N., Malignant transformation by a eukaryotic initiation factor subunit that binds to mRNA 5′ cap. Nature 345, 544–547 (1990). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

