Distinct tumor architectures and microenvironments for the initiation of breast cancer metastasis in the brain
- PMID: 39270646
- PMCID: PMC12093277
- DOI: 10.1016/j.ccell.2024.08.015
Distinct tumor architectures and microenvironments for the initiation of breast cancer metastasis in the brain
Abstract
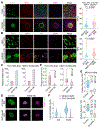
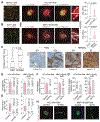
Brain metastasis, a serious complication of cancer, hinges on the initial survival, microenvironment adaptation, and outgrowth of disseminated cancer cells. To understand the early stages of brain colonization, we investigated two prevalent sources of cerebral relapse, triple-negative (TNBC) and HER2+ (HER2BC) breast cancers. Using mouse models and human tissue samples, we found that these tumor types colonize the brain, with a preference for distinctive tumor architectures, stromal interfaces, and autocrine programs. TNBC models tend to form perivascular sheaths with diffusive contact with astrocytes and microglia. In contrast, HER2BC models tend to form compact spheroids driven by autonomous tenascin C production, segregating stromal cells to the periphery. Single-cell transcriptomics of the tumor microenvironment revealed that these architectures evoke differential Alzheimer's disease-associated microglia (DAM) responses and engagement of the GAS6 receptor AXL. The spatial features of the two modes of brain colonization have relevance for leveraging the stroma to treat brain metastasis.
Keywords: brain metastasis; breast cancer; extracellular matrix; microglia; tumor architecture; tumor microenvironment.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests N.S.M. has consulted for AstraZeneca. D.P. is on the scientific advisory board of Insitro. J.M. holds company stock of Scholar Rock, Inc.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

