The Fate of a Polygenic Phenotype Within the Genomic Landscapes of Introgression in the European Seabass Hybrid Zone
- PMID: 39271153
- PMCID: PMC11430266
- DOI: 10.1093/molbev/msae194
The Fate of a Polygenic Phenotype Within the Genomic Landscapes of Introgression in the European Seabass Hybrid Zone
Abstract
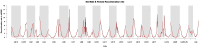
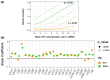
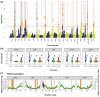
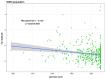
Unraveling the evolutionary mechanisms and consequences of hybridization is a major concern in biology. Many studies have documented the interplay between recombination and selection in modulating the genomic landscape of introgression, but few have considered how associations with phenotype may affect this landscape. Here, we use the European seabass (Dicentrarchus labrax), a key species in marine aquaculture that undergoes natural hybridization, to determine how selection on phenotype modulates the introgression landscape between Atlantic and Mediterranean lineages. We use a high-density single nucleotide polymorphism array to assess individual local ancestry along the genome and improve the mapping of muscle fat content, a polygenic trait that is divergent between lineages. Taking into account variation in recombination rates, we reveal a purging of Atlantic ancestry in the admixed Mediterranean populations. While Atlantic individuals had higher muscle fat content, we observed that genomic regions associated with this trait in Mediterranean populations displayed reduced introgression of Atlantic ancestry. These results emphasize how selection against maladapted alleles shapes the genomic landscape of introgression.
Keywords: European seabass; hybridization; introgression; phenotype; recombination; selection.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Conflict of interest statement
Conflict of Interest The authors declare that there is no conflict of interest.
Figures


References
-
- Anderson E, Stebbins GL. Hybridization as an evolutionary stimulus. Evolution. 1954:8(4):378–388. 10.1111/j.1558-5646.1954.tb01504.x. - DOI
-
- Atkinson EG, Maihofer AX, Kanai M, Martin AR, Karczewski KJ, Santoro ML, Ulirsch JC, Kamatani Y, Okada Y, Finucane HK, et al. Tractor uses local ancestry to enable the inclusion of admixed individuals in GWAS and to boost power. Nat Genet. 2021:53(2):195–204. 10.1038/s41588-020-00766-y. - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

