Enhanced detection of RNA modifications and read mapping with high-accuracy nanopore RNA basecalling models
- PMID: 39271295
- PMCID: PMC11610583
- DOI: 10.1101/gr.278849.123
Enhanced detection of RNA modifications and read mapping with high-accuracy nanopore RNA basecalling models
Abstract
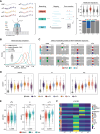
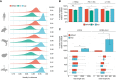
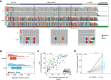
In recent years, nanopore direct RNA sequencing (DRS) became a valuable tool for studying the epitranscriptome, owing to its ability to detect multiple modifications within the same full-length native RNA molecules. Although RNA modifications can be identified in the form of systematic basecalling "errors" in DRS data sets, N6-methyladenosine (m6A) modifications produce relatively low "errors" compared with other RNA modifications, limiting the applicability of this approach to m6A sites that are modified at high stoichiometries. Here, we demonstrate that the use of alternative RNA basecalling models, trained with fully unmodified sequences, increases the "error" signal of m6A, leading to enhanced detection and improved sensitivity even at low stoichiometries. Moreover, we find that high-accuracy alternative RNA basecalling models can show up to 97% median basecalling accuracy, outperforming currently available RNA basecalling models, which show 91% median basecalling accuracy. Notably, the use of high-accuracy basecalling models is accompanied by a significant increase in the number of mapped reads-especially in shorter RNA fractions-and increased basecalling error signatures at pseudouridine (Ψ)- and N1-methylpseudouridine (m1Ψ)-modified sites. Overall, our work demonstrates that alternative RNA basecalling models can be used to improve the detection of RNA modifications, read mappability, and basecalling accuracy in nanopore DRS data sets.
© 2024 Diensthuber et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Acera Mateos P, Sethi AJ, Ravindran A, Srivastava A, Woodward K, Mahmud S, Kanchi M, Guarnacci M, Xu J, Yuen ZWS, et al. 2024. Prediction of m6A and m5C at single-molecule resolution reveals a transcriptome-wide co-occurrence of RNA modifications. Nat Commun 15: 3899. 10.1038/s41467-024-47953-7 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
