Unbiased screening identifies regulators of cell-cell adhesion and treatment options in pemphigus
- PMID: 39271654
- PMCID: PMC11399147
- DOI: 10.1038/s41467-024-51747-2
Unbiased screening identifies regulators of cell-cell adhesion and treatment options in pemphigus
Abstract
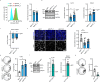
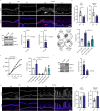
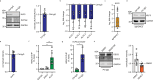
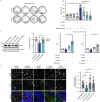
Cell-cell junctions, and specifically desmosomes, are crucial for robust intercellular adhesion. Desmosomal function is compromised in the autoimmune blistering skin disease pemphigus vulgaris. We combine whole-genome knockout screening and a promotor screen of the desmosomal gene desmoglein 3 in human keratinocytes to identify novel regulators of intercellular adhesion. Kruppel-like-factor 5 (KLF5) directly binds to the desmoglein 3 regulatory region and promotes adhesion. Reduced levels of KLF5 in patient tissue indicate a role in pemphigus vulgaris. Autoantibody fractions from patients impair intercellular adhesion and reduce KLF5 levels in in vitro and in vivo disease models. These effects were dependent on increased activity of histone deacetylase 3, leading to transcriptional repression of KLF5. Inhibiting histone deacetylase 3 increases KLF5 levels and protects against the deleterious effects of autoantibodies in murine and human pemphigus vulgaris models. Together, KLF5 and histone deacetylase 3 are regulators of desmoglein 3 gene expression and intercellular adhesion and represent potential therapeutic targets in pemphigus vulgaris.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

