Conserved transcriptional regulation by BRN1 and BRN2 in neocortical progenitors drives mammalian neural specification and neocortical expansion
- PMID: 39271675
- PMCID: PMC11399407
- DOI: 10.1038/s41467-024-52443-x
Conserved transcriptional regulation by BRN1 and BRN2 in neocortical progenitors drives mammalian neural specification and neocortical expansion
Abstract
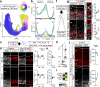
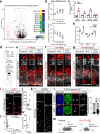
The neocortex varies in size and complexity among mammals due to the tremendous variability in the number and diversity of neuronal subtypes across species. The increased cellular diversity is paralleled by the expansion of the pool of neocortical progenitors and the emergence of indirect neurogenesis during brain evolution. The molecular pathways that control these biological processes and are disrupted in neurological disorders remain largely unknown. Here we show that the transcription factors BRN1 and BRN2 have an evolutionary conserved function in neocortical progenitors to control their proliferative capacity and the switch from direct to indirect neurogenesis. Functional studies in mice and ferrets show that BRN1/2 act in concert with NOTCH and primary microcephaly genes to regulate progenitor behavior. Analysis of transcriptomics data from genetically modified macaques provides evidence that these molecular pathways are conserved in non-human primates. Our findings thus demonstrate that BRN1/2 are central regulators of gene expression programs in neocortical progenitors critical to determine brain size during evolution.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- SRA/SRP366952
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

