HIV-1 adapts to lost IP6 coordination through second-site mutations that restore conical capsid assembly
- PMID: 39271696
- PMCID: PMC11399258
- DOI: 10.1038/s41467-024-51971-w
HIV-1 adapts to lost IP6 coordination through second-site mutations that restore conical capsid assembly
Abstract
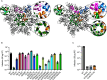
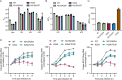
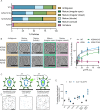
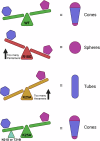
The HIV-1 capsid is composed of capsid (CA) protein hexamers and pentamers (capsomers) that contain a central pore hypothesised to regulate capsid assembly and facilitate nucleotide import early during post-infection. These pore functions are mediated by two positively charged rings created by CA Arg-18 (R18) and Lys-25 (K25). Here we describe the forced evolution of viruses containing mutations in R18 and K25. Whilst R18 mutants fail to replicate, K25A viruses acquire compensating mutations that restore nearly wild-type replication fitness. These compensating mutations, which rescue reverse transcription and infection without reintroducing lost pore charges, map to three adaptation hot-spots located within and between capsomers. The second-site suppressor mutations act by restoring the formation of pentamers lost upon K25 mutation, enabling closed conical capsid assembly both in vitro and inside virions. These results indicate that there is no intrinsic requirement for K25 in either nucleotide import or capsid assembly. We propose that whilst HIV-1 must maintain a precise hexamer:pentamer equilibrium for proper capsid assembly, compensatory mutations can tune this equilibrium to restore fitness lost by mutation of the central pore.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

