Ligand requirements for immunoreceptor triggering
- PMID: 39271744
- PMCID: PMC11399299
- DOI: 10.1038/s42003-024-06817-y
Ligand requirements for immunoreceptor triggering
Abstract
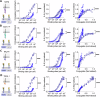
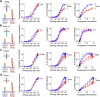
Leukocytes interact with other cells using cell surface receptors. The largest group of such receptors are non-catalytic tyrosine phosphorylated receptors (NTRs), also called immunoreceptors. NTR signalling requires phosphorylation of cytoplasmic tyrosine residues by SRC-family tyrosine kinases. How ligand binding to NTRs induces this phosphorylation, also called NTR triggering, remains controversial, with roles suggested for size-based segregation, clustering, and mechanical force. Here we exploit a recently developed cell-surface generic ligand system to explore the ligand requirements for NTR triggering. We examine the effect of varying the ligand's length, mobility and valency on the activation of representative members of four NTR families: SIRPβ1, Siglec 14, NKp44 and TREM-1. Increasing the ligand length impairs activation via NTRs, despite enhancing cell-cell conjugation, while varying ligand mobility has little effect on either conjugation or activation. Increasing the valency of the ligand, while enhancing cell-cell conjugation, does not enhance activation at equivalent levels of conjugation. These findings are more consistent with a role for size-based segregation, rather than mechanical force or clustering, in NTR triggering, suggesting a role for the kinetic-segregation model.
© 2024. The Author(s).
Conflict of interest statement
PAvdM is a founder and consultant for MatchBio Limited.
Figures



References
-
- Barclay, A. N. et al. The Leucocyte Antigen Factsbook. (Academic Press, 1997).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

