Integrative gene regulatory network analysis discloses key driver genes of fibromuscular dysplasia
- PMID: 39271816
- PMCID: PMC12413977
- DOI: 10.1038/s44161-024-00533-w
Integrative gene regulatory network analysis discloses key driver genes of fibromuscular dysplasia
Abstract
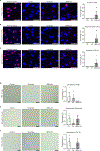
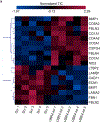
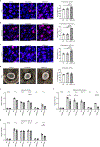
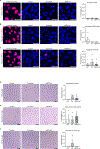
Fibromuscular dysplasia (FMD) is a poorly understood disease affecting 3-5% of adult females. The pathobiology of FMD involves arterial lesions of stenosis, dissection, tortuosity, dilation and aneurysm, which can lead to hypertension, stroke, myocardial infarction and even death. Currently, there are no animal models for FMD and few insights as to its pathobiology. In this study, by integrating DNA genotype and RNA sequence data from primary fibroblasts of 83 patients with FMD and 71 matched healthy controls, we inferred 18 gene regulatory co-expression networks, four of which were found to act together as an FMD-associated supernetwork in the arterial wall. After in vivo perturbation of this co-expression supernetwork by selective knockout of a top network key driver, mice developed arterial dilation, a hallmark of FMD. Molecular studies indicated that this supernetwork governs multiple aspects of vascular cell physiology and functionality, including collagen/matrix production. These studies illuminate the complex causal mechanisms of FMD and suggest a potential therapeutic avenue for this challenging disease.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests Statement
The other authors have no conflicts of interest or relationships with industry to declare.
Figures











References
-
- Sottiurai VS, Fry WJ & Stanley JC Ultrastructure of medial smooth muscle and myofibroblasts in human arterial dysplasia. Arch Surg 113, 1280–8 (1978). - PubMed
-
- Olin JW et al. The United States Registry for Fibromuscular Dysplasia: results in the first 447 patients. Circulation 125, 3182–90 (2012). - PubMed
-
- Pappaccogli M et al. The European/International Fibromuscular Dysplasia Registry and Initiative (FEIRI)-clinical phenotypes and their predictors based on a cohort of 1000 patients. Cardiovasc Res 117, 950–959 (2021). - PubMed
-
- Gornik HL et al. First International Consensus on the diagnosis and management of fibromuscular dysplasia. Vasc Med 24, 164–189 (2019). - PubMed
-
- Hendricks NJ et al. Is fibromuscular dysplasia underdiagnosed? A comparison of the prevalence of FMD seen in CORAL trial participants versus a single institution population of renal donor candidates. Vasc Med 19, 363–7 (2014). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
