Tumour immune characterisation of primary triple-negative breast cancer using automated image quantification of immunohistochemistry-stained immune cells
- PMID: 39271910
- PMCID: PMC11399404
- DOI: 10.1038/s41598-024-72306-1
Tumour immune characterisation of primary triple-negative breast cancer using automated image quantification of immunohistochemistry-stained immune cells
Abstract
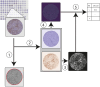
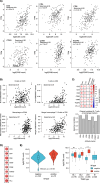
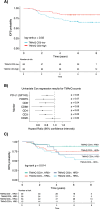
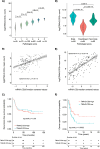
The tumour immune microenvironment (TIME) in breast cancer is acknowledged with an increasing role in treatment response and prognosis. With a growing number of immune markers analysed, digital image analysis may facilitate broader TIME understanding, even in single-plex IHC data. To facilitate analyses of the latter an open-source image analysis pipeline, Tissue microarray MArker Quantification (TMArQ), was developed and applied to single-plex stainings for p53, CD3, CD4, CD8, CD20, CD68, FOXP3, and PD-L1 (SP142 antibody) in a 218-patient triple negative breast cancer (TNBC) cohort with complementary pathology scorings, clinicopathological, whole genome sequencing, and RNA-sequencing data. TMArQ's cell counts for analysed immune markers were on par with results from alternative methods and consistent with both estimates from human pathology review, different quantifications and classifications derived from RNA-sequencing as well as known prognostic patterns of immune response in TNBC. The digital cell counts demonstrated how immune markers are coexpressed in the TIME when considering TNBC molecular subtypes and DNA repair deficiency, and how combination of immune status with DNA repair deficiency status can improve the prognostic stratification in chemotherapy treated patients. These results underscore the value and potential of integrating TIME and specific tumour intrinsic alterations/phenotypes for the molecular understanding of TNBC.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- de Jong, V. M. T. et al. Prognostic value of stromal tumor-infiltrating lymphocytes in young, node-negative, triple-negative breast cancer patients who did not receive (neo)adjuvant systemic therapy. J. Clin. Oncol.40, 2361–2374. 10.1200/JCO.21.01536 (2022). 10.1200/JCO.21.01536 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

