Single-cell, single-nucleus and xenium-based spatial transcriptomics analyses reveal inflammatory activation and altered cell interactions in the hippocampus in mice with temporal lobe epilepsy
- PMID: 39272194
- PMCID: PMC11396644
- DOI: 10.1186/s40364-024-00636-3
Single-cell, single-nucleus and xenium-based spatial transcriptomics analyses reveal inflammatory activation and altered cell interactions in the hippocampus in mice with temporal lobe epilepsy
Abstract
Background: Temporal lobe epilepsy (TLE) is among the most common types of epilepsy and often leads to cognitive, emotional, and psychiatric issues due to the frequent seizures. A notable pathological change related to TLE is hippocampal sclerosis (HS), which is characterized by neuronal loss, gliosis, and an increased neuron fibre density. The mechanisms underlying TLE-HS development remain unclear, but the reactive transcriptomic changes in glial cells and neurons of the hippocampus post-epileptogenesis may provide insights.
Methods: To induce TLE, 200 nl of kainic acid (KA) was stereotactically injected into the hippocampal CA1 region of mice, followed by a 7-day postinjection period. Single-cell RNA sequencing (ScRNA-seq), single-nucleus RNA sequencing (SnRNA-seq), and Xenium-based spatial transcriptomics analyses were employed to evaluate the changes in mRNA expression in glial cells and neurons.
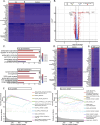
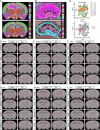
Results: From the ScRNA-seq and SnRNA-seq data, 31,390 glial cells and 48,221 neuronal nuclei were identified. Analysis of the differentially expressed genes (DEGs) revealed significant transcriptomic alterations in the hippocampal cells of mice with TLE, affecting hundreds to thousands of mRNAs and their signalling pathways. Enrichment analysis indicated notable activation of stress and inflammatory pathways in the TLE hippocampus, while pathways related to axonal development and neural support were suppressed. Xenium analysis demonstrated the expression of all 247 genes across mouse brain sections, revealing the spatial distributions of their expression in 27 cell types. Integrated analysis of the DEGs identified via the three sequencing techniques revealed that Spp1, Trem2, and Cd68 were upregulated in all glial cell types and in the Xenium data; Penk, Sorcs3, and Plekha2 were upregulated in all neuronal cell types and in the Xenium data; and Tle4 and Sipa1l3 were downregulated in all glial cell types and in the Xenium data.
Conclusion: In this study, a high-resolution single-cell transcriptomic atlas of the hippocampus in mice with TLE was established, revealing potential intrinsic mechanisms driving TLE-associated inflammatory activation and altered cell interactions. These findings provide valuable insights for further exploration of HS development and epileptogenesis.
Keywords: Single-cell RNA sequencing; Single-nucleus RNA sequencing; Temporal lobe epilepsy; Xenium-based spatial transcriptomics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

