A triple hormone receptor ER, AR, and VDR signature is a robust prognosis predictor in breast cancer
- PMID: 39272208
- PMCID: PMC11395215
- DOI: 10.1186/s13058-024-01876-9
A triple hormone receptor ER, AR, and VDR signature is a robust prognosis predictor in breast cancer
Abstract
Background: Despite evidence indicating the dominance of cell-of-origin signatures in molecular tumor patterns, translating these genome-wide patterns into actionable insights has been challenging. This study introduces breast cancer cell-of-origin signatures that offer significant prognostic value across all breast cancer subtypes and various clinical cohorts, compared to previously developed genomic signatures.
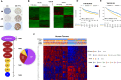
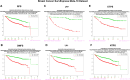
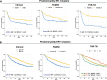
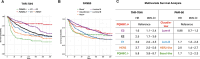
Methods: We previously reported that triple hormone receptor (THR) co-expression patterns of androgen (AR), estrogen (ER), and vitamin D (VDR) receptors are maintained at the protein level in human breast cancers. Here, we developed corresponding mRNA signatures (THR-50 and THR-70) based on these patterns to categorize breast tumors by their THR expression levels. The THR mRNA signatures were evaluated across 56 breast cancer datasets (5040 patients) using Kaplan-Meier survival analysis, Cox proportional hazard regression, and unsupervised clustering.
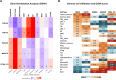
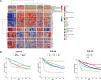
Results: The THR signatures effectively predict both overall and progression-free survival across all evaluated datasets, independent of subtype, grade, or treatment status, suggesting improvement over existing prognostic signatures. Furthermore, they delineate three distinct ER-positive breast cancer subtypes with significant survival in differences-expanding on the conventional two subtypes. Additionally, coupling THR-70 with an immune signature identifies a predominantly ER-negative breast cancer subgroup with a highly favorable prognosis, comparable to ER-positive cases, as well as an ER-negative subgroup with notably poor outcome, characterized by a 15-fold shorter survival.
Conclusions: The THR cell-of-origin signature introduces a novel dimension to breast cancer biology, potentially serving as a robust foundation for integrating additional prognostic biomarkers. These signatures offer utility as a prognostic index for stratifying existing breast cancer subtypes and for de novo classification of breast cancer cases. Moreover, THR signatures may also hold promise in predicting hormone treatment responses targeting AR and/or VDR.
Keywords: Androgen receptor; Breast cancer; Breast cancer subtypes; Cell-of-origin; Estrogen receptor; Predictive modeling; Survival; Triple hormone receptor; Vitamin D receptor.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, Bejar R, Berti E, Busque L, Chan JK, Chen W. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36(7):1703–19. 10.1038/s41375-022-01613-1 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

