RNA-Seq Analysis of Glycolysis Regulation of Avian Leukosis Virus Subgroup J Replication
- PMID: 39272286
- PMCID: PMC11394362
- DOI: 10.3390/ani14172500
RNA-Seq Analysis of Glycolysis Regulation of Avian Leukosis Virus Subgroup J Replication
Abstract
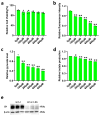
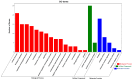
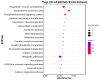
Avian Leukosis virus (ALV) is a widely spread virus that causes major economic losses to the global poultry industry. This study aims to investigate the effect of glycolysis on the replication of the ALV-J virus and identify the key circular RNAs that regulate the replication of the ALV-J virus. We found that glucose uptake, pyruvate content, and lactate content in DF1 cells were increased after ALV-J infection. Moreover, inhibiting the glycolysis of ALV-J-infected DF1 cells reduced the replication of the ALV-J virus. To further study the mechanism of glycolysis in the replication of the ALV-J virus, we performed RNA-seq on ALV-J-infected and ALV-J-infected cells treated with glycolysis inhibition. RNA-seq results show that a total of 10,375 circular RNAs (circRNAs) were identified, of which the main types were exonic circular RNAs, and 28 circRNAs were differentially expressed between ALV-J-infected and ALV-J-infected cells treated with glycolysis inhibition. Then, we performed functional enrichment analysis of differentially expressed circRNA source and target genes. Functional enrichment analysis indicated that some circRNAs might be involved in regulating the replication of the ALV-J virus by influencing some pathways like glycolysis/gluconeogenesis, the NOD-like receptor signaling pathway, MAPK signaling pathway, p53 signaling pathway, Toll-like receptor signaling pathway, Insulin signaling pathway, and Apoptosis. This study revealed the effect of glycolysis on the replication of the ALV-J virus in DF1 cells and its possible regulatory mechanism, which provided a basis for understanding the factors influencing the replication of the ALV-J virus and reducing the rate of infection of the ALV-J virus in poultry.
Keywords: ALV-J; CircRNA; glycolysis; virus replication.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Li X., Lin W., Chang S., Zhao P., Zhang X., Liu Y., Chen W., Li B., Shu D., Zhang H. Isolation, identification and evolution analysis of a novel subgroup of avian leukosis virus isolated from a local Chinese yellow broiler in South China. Arch. Virol. 2016;161:2717–2725. doi: 10.1007/s00705-016-2965-x. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

