Genome-Wide CRISPR Screen Identifies KEAP1 Perturbation as a Vulnerability of ARID1A-Deficient Cells
- PMID: 39272807
- PMCID: PMC11394604
- DOI: 10.3390/cancers16172949
Genome-Wide CRISPR Screen Identifies KEAP1 Perturbation as a Vulnerability of ARID1A-Deficient Cells
Abstract
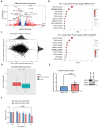
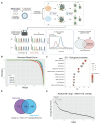
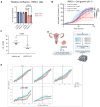
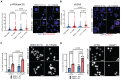
ARID1A is the core DNA-binding subunit of the BAF chromatin remodeling complex and is mutated in about 8% of all cancers. The frequency of ARID1A loss varies between cancer subtypes, with clear cell ovarian carcinoma (CCOC) presenting the highest incidence at > 50% of cases. Despite a growing understanding of the consequences of ARID1A loss in cancer, there remains limited targeted therapeutic options for ARID1A-deficient cancers. Using a genome-wide CRISPR screening approach, we identify KEAP1 as a genetic dependency of ARID1A in CCOC. Depletion or chemical perturbation of KEAP1 results in selective growth inhibition of ARID1A-KO cell lines and edited primary endometrial epithelial cells. While we confirm that KEAP1-NRF2 signalling is dysregulated in ARID1A-KO cells, we suggest that this synthetic lethality is not due to aberrant NRF2 signalling. Rather, we find that KEAP1 perturbation exacerbates genome instability phenotypes associated with ARID1A deficiency. Together, our findings identify a potentially novel synthetic lethal interaction of ARID1A-deficient cells.
Keywords: ARID1A; CRISPR screening; KEAP1; synthetic lethality.
Conflict of interest statement
LAF is a member of the Office of the Canada Chief Science Advisor Youth Council (CSA-YC). PCS is CEO of Arrowsmith Genetics Inc.
Figures
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

