A Comparison of Two Versions of the CRISPR-Sirius System for the Live-Cell Visualization of the Borders of Topologically Associating Domains
- PMID: 39273012
- PMCID: PMC11394217
- DOI: 10.3390/cells13171440
A Comparison of Two Versions of the CRISPR-Sirius System for the Live-Cell Visualization of the Borders of Topologically Associating Domains
Abstract
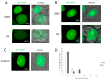
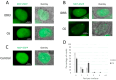
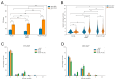
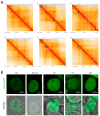
In recent years, various technologies have emerged for the imaging of chromatin loci in living cells via catalytically inactive Cas9 (dCas9). These technologies facilitate a deeper understanding of the mechanisms behind the chromatin dynamics and provide valuable kinetic data that could not have previously been obtained via FISH applied to fixed cells. However, such technologies are relatively complicated, as they involve the expression of several chimeric proteins as well as sgRNAs targeting the visualized loci, a process that entails many technical subtleties. Therefore, the effectiveness in visualizing a specific target locus may be quite low. In this study, we directly compared two versions of a previously published CRISPR-Sirius method based on the use of sgRNAs containing eight MS2 or PP7 stem loops and the expression of MCP or PCP fused to fluorescent proteins. We assessed the visualization efficiency for several unique genomic loci by comparing the two approaches in delivering sgRNA genes (transient transfection and lentiviral transduction), as well as two CRISPR-Sirius versions (with PCP and with MCP). The efficiency of visualization varied among the loci, and not all loci could be visualized. However, the MCP-sfGFP version provided more efficient visualization in terms of the number of cells with signals than PCP-sfGFP for all tested loci. We also showed that lentiviral transduction was more efficient in locus imaging than transient transfection for both CRISPR-Sirius systems. Most of the target loci in our study were located at the borders of topologically associating domains, and we defined a set of TAD borders that could be effectively visualized using the MCP-sfGFP version of the CRISPR-Sirius system. Altogether, our study validates the use of the CRISPR-Sirius technology for live-cell visualization and highlights various technical details that should be considered when using this method.
Keywords: 4D genome; CRISPR-Sirius; CRISPR-imaging; chromatin visualization; cohesin; live-cell microscopy; topologically associating domains (TADs).
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of the data; in the writing of the manuscript; or in the decision to publish the results.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

