Functional Insights in PLS3-Mediated Osteogenic Regulation
- PMID: 39273077
- PMCID: PMC11394082
- DOI: 10.3390/cells13171507
Functional Insights in PLS3-Mediated Osteogenic Regulation
Abstract
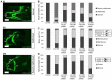
Plastin-3 (PLS3) encodes T-plastin, an actin-bundling protein mediating the formation of actin filaments by which numerous cellular processes are regulated. Loss-of-function genetic defects in PLS3 are reported to cause X-linked osteoporosis and childhood-onset fractures. However, the molecular etiology of PLS3 remains elusive. Functional compensation by actin-bundling proteins ACTN1, ACTN4, and FSCN1 was investigated in zebrafish following morpholino-mediated pls3 knockdown. Primary dermal fibroblasts from six patients with a PLS3 variant were also used to examine expression of these proteins during osteogenic differentiation. In addition, Pls3 knockdown in the murine MLO-Y4 cell line was employed to provide insights in global gene expression. Our results showed that ACTN1 and ACTN4 can rescue the skeletal deformities in zebrafish after pls3 knockdown, but this was inadequate for FSCN1. Patients' fibroblasts showed the same osteogenic transdifferentiation ability as healthy donors. RNA-seq results showed differential expression in Wnt1, Nos1ap, and Myh3 after Pls3 knockdown in MLO-Y4 cells, which were also associated with the Wnt and Th17 cell differentiation pathways. Moreover, WNT2 was significantly increased in patient osteoblast-like cells compared to healthy donors. Altogether, our findings in different bone cell types indicate that the mechanism of PLS3-related pathology extends beyond actin-bundling proteins, implicating broader pathways of bone metabolism.
Keywords: actin-bundling protein; osteogenic differentiation; osteoporosis; plastin-3; transcriptome.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

