Satellitome Analysis of Adalia bipunctata (Coleoptera): Revealing Centromeric Turnover and Potential Chromosome Rearrangements in a Comparative Interspecific Study
- PMID: 39273162
- PMCID: PMC11394905
- DOI: 10.3390/ijms25179214
Satellitome Analysis of Adalia bipunctata (Coleoptera): Revealing Centromeric Turnover and Potential Chromosome Rearrangements in a Comparative Interspecific Study
Abstract
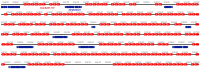
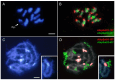
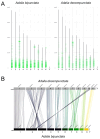
Eukaryotic genomes exhibit a dynamic interplay between single-copy sequences and repetitive DNA elements, with satellite DNA (satDNA) representing a substantial portion, mainly situated at telomeric and centromeric chromosomal regions. We utilized Illumina next-generation sequencing data from Adalia bipunctata to investigate its satellitome. Cytogenetic mapping via fluorescence in situ hybridization was performed for the most abundant satDNA families. In silico localization of satDNAs was carried out using the CHRISMAPP (Chromosome In Silico Mapping) pipeline on the high-fidelity chromosome-level assembly already available for this species, enabling a meticulous characterization and localization of multiple satDNA families. Additionally, we analyzed the conservation of the satellitome at an interspecific scale. Specifically, we employed the CHRISMAPP pipeline to map the satDNAs of A. bipunctata onto the genome of Adalia decempunctata, which has also been sequenced and assembled at the chromosome level. This analysis, along with the creation of a synteny map between the two species, suggests a rapid turnover of centromeric satDNA between these species and the potential occurrence of chromosomal rearrangements, despite the considerable conservation of their satellitomes. Specific satDNA families in the sex chromosomes of both species suggest a role in sex chromosome differentiation. Our interspecific comparative study can provide a significant advance in the understanding of the repeat genome organization and evolution in beetles.
Keywords: Adalia bipunctata; Adalia decempunctata; Chromosome In Silico Mapping (CHRISMAPP); Coccinellidae; Coleoptera; RepeatExplorer2; fluorescence in situ hybridization (FISH); karyotype evolution; ladybird beetle; satellite DNA; satellitome.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

