Genetic Diversity, Selection Signatures, and Genome-Wide Association Study Identify Candidate Genes Related to Litter Size in Hu Sheep
- PMID: 39273345
- PMCID: PMC11395453
- DOI: 10.3390/ijms25179397
Genetic Diversity, Selection Signatures, and Genome-Wide Association Study Identify Candidate Genes Related to Litter Size in Hu Sheep
Abstract
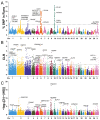
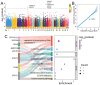
Hu sheep is a renowned prolific local sheep breed in China, widely distributed across the country due to its excellent reproductive performance. Deciphering the molecular mechanisms underlying the high fecundity of Hu sheep is crucial for improving the litter size of ewes. In this study, we genotyped 830 female Hu sheep using the Illumina OvineSNP50 BeadChip and performed genetic diversity analysis, selection signature detection, and a genome-wide association study (GWAS) for litter size. Our results revealed that the Hu sheep population exhibits relatively high genetic diversity. A total of 4927 runs of homozygosity (ROH) segments were detected, with the majority (74.73%) being short in length. Different genomic inbreeding coefficients (FROH, FHOM, FGRM, and FUNI) ranged from -0.0060 to 0.0126, showing low levels of inbreeding in this population. Additionally, we identified 91 candidate genomic regions through three complementary selection signature methods, including ROH, composite likelihood ratio (CLR), and integrated haplotype score (iHS), and annotated 189 protein-coding genes. Moreover, we observed two significant SNPs related to the litter size of Hu sheep using GWAS analysis based on a repeatability model. Integrating the selection signatures and the GWAS results, we identified 15 candidate genes associated with litter size, among which BMPR1B and UNC5C were particularly noteworthy. These findings provide valuable insights for improving the reproductive performance and breeding of high-fecundity lines of Hu sheep.
Keywords: GWAS; Hu sheep; candidate genes; genetic diversity; litter size; selection signatures.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Amer P.R., McEwan J.C., Dodds K.G., Davis G.H. Economic values for ewe prolificacy and lamb survival in New Zealand sheep. Livest. Prod. Sci. 1999;58:75–90. doi: 10.1016/S0301-6226(98)00192-4. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

