Long-Read MDM4 Sequencing Reveals Aberrant Isoform Landscape in Metastatic Melanomas
- PMID: 39273363
- PMCID: PMC11395681
- DOI: 10.3390/ijms25179415
Long-Read MDM4 Sequencing Reveals Aberrant Isoform Landscape in Metastatic Melanomas
Abstract
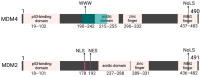
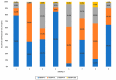
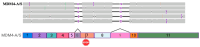
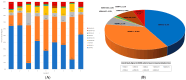
MDM4 is upregulated in the majority of melanoma cases and has been described as a "key therapeutic target in cutaneous melanoma". Numerous isoforms of MDM4 exist, with few studies examining their specific expression in human tissues. The changes in splicing of MDM4 during human melanomagenesis are critical to p53 activity and represent potential therapeutic targets. Compounding this, studies relying on short reads lose "connectivity" data, so full transcripts are frequently only inferred from the presence of splice junction reads. To address this problem, long-read nanopore sequencing was utilized to read the entire length of transcripts. Here, MDM4 transcripts, both alternative and canonical, are characterized in a pilot cohort of human melanoma specimens. RT-PCR was first used to identify the presence of novel splice junctions in these specimens. RT-qPCR then quantified the expression of major MDM4 isoforms observed during sequencing. The current study both identifies and quantifies MDM4 isoforms present in melanoma tumor samples. In the current study, we observed high expression levels of MDM4-S, MDM4-FL, MDM4-A, and the previously undescribed Ensembl transcript MDM4-209. A novel transcript lacking both exons 6 and 9 is observed and named MDM4-A/S for its resemblance to both MDM4-A and MDM4-S isoforms.
Keywords: MDM4; alternative splicing; isoforms; long-read nanopore sequencing; melanoma.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





References
-
- Artosi F., Costanza G., Di Prete M., Garofalo V., Lozzi F., Dika E., Cosio T., Diluvio L., Shumak R.G., Lambiase S., et al. Epidemiological and Clinical Analysis of Exposure-Related Factors in Non-Melanoma Skin Cancer: A Retrospective Cohort Study. Environ. Res. 2024;247:118117. doi: 10.1016/j.envres.2024.118117. - DOI - PubMed
-
- American Cancer Society Melanoma Skin Cancer Statistics. [(accessed on 27 February 2024)]. Available online: https://www.cancer.org/cancer/types/melanoma-skin-cancer/about/key-stati....
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

