Specific microRNA Profile Associated with Inflammation and Lipid Metabolism for Stratifying Allergic Asthma Severity
- PMID: 39273372
- PMCID: PMC11394998
- DOI: 10.3390/ijms25179425
Specific microRNA Profile Associated with Inflammation and Lipid Metabolism for Stratifying Allergic Asthma Severity
Abstract
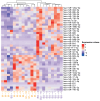
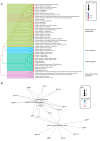
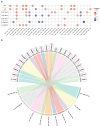
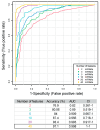
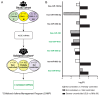
The mechanisms underlying severe allergic asthma are complex and unknown, meaning it is a challenge to provide the most appropriate treatment. This study aimed to identify novel biomarkers for stratifying allergic asthmatic patients according to severity, and to uncover the biological mechanisms that lead to the development of the severe uncontrolled phenotype. By using miRNA PCR panels, we analyzed the expression of 752 miRNAs in serum samples from control subjects (n = 15) and mild (n = 11) and severe uncontrolled (n = 10) allergic asthmatic patients. We identified 40 differentially expressed miRNAs between severe uncontrolled and mild allergic asthmatic patients. Functional enrichment analysis revealed signatures related to inflammation, angiogenesis, lipid metabolism and mRNA regulation. A random forest classifier trained with DE miRNAs achieved a high accuracy of 97% for severe uncontrolled patient stratification. Validation of the identified biomarkers was performed on a subset of allergic asthmatic patients from the CAMP cohort at Brigham and Women's Hospital, Harvard Medical School. Four of these miRNAs (hsa-miR-99b-5p, hsa-miR-451a, hsa-miR-326 and hsa-miR-505-3p) were validated, pointing towards their potential as biomarkers for stratifying allergic asthmatic patients by severity and providing insights into severe uncontrolled asthma molecular pathways.
Keywords: allergic asthma; biomarkers; miRNAs; severity.
Conflict of interest statement
CT reports conflicts of interest in terms of consulting fees by the GLG group and payment or honoraria for lectures, presentations, speakers’ bureaus, manuscript writing or educational events by San Pablo CEU University and Genotipia. MME reports conflicts of interest in terms of the funding for the present manuscript by RICORS Red de Enfermedades Inflamatorias (REI) RD21 0002 0008 and by Fundación Mutua Madrileña (AP177712021) and a leadership or fiduciary role in EAACI EXCOM. MJM reports conflicts of interest in terms of the funding for the present manuscript by NIH NHLBI Grants. MIDD reports conflicts of interest in terms of grants or contracts from ALK. KGT reports conflicts of interest in terms of the funding for the present manuscript by the National Institutes of Health. DB reports conflicts of interest in terms of funding for the present manuscript by ISCIII (PI19/00044) (co-founded by FERDER); grants or contracts from ALK A/S; and payment or honoraria for lectures, presentations, speakers’ bureaus, manuscript writing or educational events by Diater Labs. The rest of the authors have no conflicts of interest.
Figures
References
-
- World Health Organization Asthma. [(accessed on 7 August 2024)]; Available online: https://www.who.int/news-room/fact-sheets/detail/asthma#:~:text=Asthma%2....
-
- Wang Z., Li Y., Gao Y., Fu Y., Lin J., Lei X., Zheng J., Jiang M. Global, Regional, and National Burden of Asthma and Its Attributable Risk Factors from 1990 to 2019: A Systematic Analysis for the Global Burden of Disease Study 2019. Respir. Res. 2023;24:169. doi: 10.1186/s12931-023-02475-6. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

