Spatiotemporal Dysregulation of Neuron-Glia Related Genes and Pro-/Anti-Inflammatory miRNAs in the 5xFAD Mouse Model of Alzheimer's Disease
- PMID: 39273422
- PMCID: PMC11394861
- DOI: 10.3390/ijms25179475
Spatiotemporal Dysregulation of Neuron-Glia Related Genes and Pro-/Anti-Inflammatory miRNAs in the 5xFAD Mouse Model of Alzheimer's Disease
Abstract
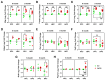
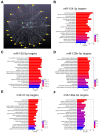
Alzheimer's disease (AD), the leading cause of dementia, is a multifactorial disease influenced by aging, genetics, and environmental factors. miRNAs are crucial regulators of gene expression and play significant roles in AD onset and progression. This exploratory study analyzed the expression levels of 28 genes and 5 miRNAs (miR-124-3p, miR-125b-5p, miR-21-5p, miR-146a-5p, and miR-155-5p) related to AD pathology and neuroimmune responses using RT-qPCR. Analyses were conducted in the prefrontal cortex (PFC) and the hippocampus (HPC) of the 5xFAD mouse AD model at 6 and 9 months old. Data highlighted upregulated genes encoding for glial fibrillary acidic protein (Gfap), triggering receptor expressed on myeloid cells (Trem2) and cystatin F (Cst7), in the 5xFAD mice at both regions and ages highlighting their roles as critical disease players and potential biomarkers. Overexpression of genes encoding for CCAAT enhancer-binding protein alpha (Cebpa) and myelin proteolipid protein (Plp) in the PFC, as well as for BCL2 apoptosis regulator (Bcl2) and purinergic receptor P2Y12 (P2yr12) in the HPC, together with upregulated microRNA(miR)-146a-5p in the PFC, prevailed in 9-month-old animals. miR-155 positively correlated with miR-146a and miR-21 in the PFC, and miR-125b positively correlated with miR-155, miR-21, while miR-146a in the HPC. Correlations between genes and miRNAs were dynamic, varying by genotype, region, and age, suggesting an intricate, disease-modulated interaction between miRNAs and target pathways. These findings contribute to our understanding of miRNAs as therapeutic targets for AD, given their multifaceted effects on neurons and glial cells.
Keywords: 5xFAD mouse model; Alzheimer’s disease; hippocampus; microRNAs; neurodegeneration; neuroimmune genes; neuroinflammation; prefrontal cortex.
Conflict of interest statement
The authors declare no conflicts of interest. The founders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



References
-
- McKeever P.M., Schneider R., Taghdiri F., Weichert A., Multani N., Brown R.A., Boxer A.L., Karydas A., Miller B., Robertson J., et al. MicroRNA Expression Levels Are Altered in the Cerebrospinal Fluid of Patients with Young-Onset Alzheimer’s Disease. Mol. Neurobiol. 2018;55:8826–8841. doi: 10.1007/s12035-018-1032-x. - DOI - PMC - PubMed
-
- Pittock R.R., Aakre J.A., Castillo A.M., Ramanan V.K., Kremers W.K., Jack C.R., Jr., Vemuri P., Lowe V.J., Knopman D.S., Petersen R.C., et al. Eligibility for Anti-Amyloid Treatment in a Population-Based Study of Cognitive Aging. Neurology. 2023;101:e1837–e1849. doi: 10.1212/WNL.0000000000207770. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

