A Novel Microbial Dysbiosis Index and Intestinal Microbiota-Associated Markers as Tools of Precision Medicine in Inflammatory Bowel Disease Paediatric Patients
- PMID: 39273567
- PMCID: PMC11395508
- DOI: 10.3390/ijms25179618
A Novel Microbial Dysbiosis Index and Intestinal Microbiota-Associated Markers as Tools of Precision Medicine in Inflammatory Bowel Disease Paediatric Patients
Abstract
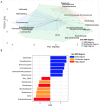
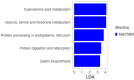
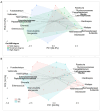
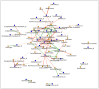
Recent evidence indicates that the gut microbiota (GM) has a significant impact on the inflammatory bowel disease (IBD) progression. Our aim was to investigate the GM profiles, the Microbial Dysbiosis Index (MDI) and the intestinal microbiota-associated markers in relation to IBD clinical characteristics and disease state. We performed 16S rRNA metataxonomy on both stools and ileal biopsies, metabolic dysbiosis tests on urine and intestinal permeability and mucosal immunity activation tests on the stools of 35 IBD paediatric patients. On the GM profile, we assigned the MDI to each patient. In the statistical analyses, the MDI was correlated with clinical parameters and intestinal microbial-associated markers. In IBD patients with high MDI, Gemellaceae and Enterobacteriaceae were increased in stools, and Fusobacterium, Haemophilus and Veillonella were increased in ileal biopsies. Ruminococcaceae and WAL_1855D were enriched in active disease condition; the last one was also positively correlated to MDI. Furthermore, the MDI results correlated with PUCAI and Matts scores in ulcerative colitis patients (UC). Finally, in our patients, we detected metabolic dysbiosis, intestinal permeability and mucosal immunity activation. In conclusion, the MDI showed a strong association with both severity and activity of IBD and a positive correlation with clinical scores, especially in UC. Thus, this evidence could be a useful tool for the diagnosis and prognosis of IBD.
Keywords: biomarkers; disease severity; gut microbiota; inflammatory bowel disease (IBD); intestinal permeability; metabolic dysbiosis; microbial dysbiosis index (MDI); mucosal immunity activation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Leylabadlo H.E., Ghotaslou R., Feizabadi M.M., Farajnia S., Moaddab S.Y., Ganbarov K., Khodadadi E., Tanomand A., Sheykhsaran E., Yousefi B., et al. The Critical Role of Faecalibacterium Prausnitzii in Human Health: An Overview. Microb. Pathog. 2020;149:104344. doi: 10.1016/j.micpath.2020.104344. - DOI - PubMed
-
- Mah C., Jayawardana T., Leong G., Koentgen S., Lemberg D., Connor S.J., Rokkas T., Grimm M.C., Leach S.T., Hold G.L. Assessing the Relationship between the Gut Microbiota and Inflammatory Bowel Disease Therapeutics: A Systematic Review. Pathogens. 2023;12:262. doi: 10.3390/pathogens12020262. - DOI - PMC - PubMed
-
- Feng C., Zhang W., Zhang T., He Q., Kwok L.-Y., Tan Y., Zhang H. Heat-Killed Bifidobacterium Bifidum B1628 May Alleviate Dextran Sulfate Sodium-Induced Colitis in Mice, and the Anti-Inflammatory Effect Is Associated with Gut Microbiota Modulation. Nutrients. 2022;14:5233. doi: 10.3390/nu14245233. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

