Exploring the Rhizospheric Microbial Communities under Long-Term Precipitation Regime in Norway Spruce Seed Orchard
- PMID: 39273604
- PMCID: PMC11395193
- DOI: 10.3390/ijms25179658
Exploring the Rhizospheric Microbial Communities under Long-Term Precipitation Regime in Norway Spruce Seed Orchard
Abstract
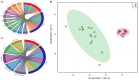
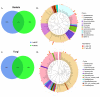
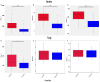
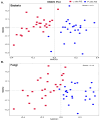
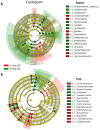
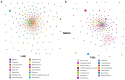
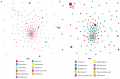
The rhizosphere is the hotspot for microbial enzyme activities and contributes to carbon cycling. Precipitation is an important component of global climate change that can profoundly alter belowground microbial communities. However, the impact of precipitation on conifer rhizospheric microbial populations has not been investigated in detail. In the present study, using high-throughput amplicon sequencing, we investigated the impact of precipitation on the rhizospheric soil microbial communities in two Norway Spruce clonal seed orchards, Lipová Lhota (L-site) and Prenet (P-site). P-site has received nearly double the precipitation than L-site for the last three decades. P-site documented higher soil water content with a significantly higher abundance of Aluminium (Al), Iron (Fe), Phosphorous (P), and Sulphur (S) than L-site. Rhizospheric soil metabolite profiling revealed an increased abundance of acids, carbohydrates, fatty acids, and alcohols in P-site. There was variance in the relative abundance of distinct microbiomes between the sites. A higher abundance of Proteobacteria, Acidobacteriota, Ascomycota, and Mortiellomycota was observed in P-site receiving high precipitation, while Bacteroidota, Actinobacteria, Chloroflexi, Firmicutes, Gemmatimonadota, and Basidiomycota were prevalent in L-site. The higher clustering coefficient of the microbial network in P-site suggested that the microbial community structure is highly interconnected and tends to cluster closely. The current study unveils the impact of precipitation variations on the spruce rhizospheric microbial association and opens new avenues for understanding the impact of global change on conifer rizospheric microbial associations.
Keywords: FUNGuild; Norway spruce; PICRUSt2; amplicon sequencing; microbial communities; network analysis; precipitation; rhizosphere; seed orchards; soil metabolites.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



References
-
- Malhi Y., Baldocchi D., Jarvis P. The carbon balance of tropical, temperate and boreal forests. Plant Cell Environ. 1999;22:715–740. doi: 10.1046/j.1365-3040.1999.00453.x. - DOI
-
- Martinović T., Mašínová T., López-Mondéjar R., Jansa J., Štursová M., Starke R., Baldrian P. Microbial utilization of simple and complex carbon compounds in a temperate forest soil. Soil Biol. Biochem. 2022;173:108786. doi: 10.1016/j.soilbio.2022.108786. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

