Variation in Root-Associated Microbial Communities among Three Different Plant Species in Natural Desert Ecosystem
- PMID: 39273952
- PMCID: PMC11396838
- DOI: 10.3390/plants13172468
Variation in Root-Associated Microbial Communities among Three Different Plant Species in Natural Desert Ecosystem
Abstract
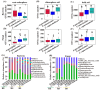
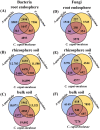
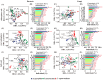
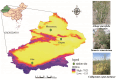
The process and function that underlie the assembly of root-associated microbiomes may be strongly linked to the survival strategy of plants. However, the assembly and functional changes of root-associated microbial communities in different desert plants in natural desert ecosystems are still unclear. Thus, we studied the microbial communities and diversity of root endosphere (RE), rhizosphere soil (RS), and bulk soil (BS) among three representative desert plants (Alhagi sparsifolia, Tamarix ramosissima, and Calligonum caput-medusae) in three Xinjiang desert regions {Taklimakan (CL), Gurbantünggüt (MSW), and Kumtag (TLF)} in China. This study found that the soil properties {electrical conductivity (EC), soil organic carbon (SOC), total nitrogen (TN) and phosphorus (TP), available nitrogen (AN) and phosphorus (AP)} of C. caput-medusae were significantly lower than those of A. sparsifolia and T. ramosissima, while the root nutrients (TN and TP) of A. sparsifolia were significantly higher compared to C. caput-medusae and T. ramosissima. The beta diversity of bacteria and fungi (RE) among the three desert plants was significantly different. The common OTU numbers of bacteria and fungi in three compartments (RE, RS, and BS) of the three desert plants were ranked as RS > BS > RE. The bacterial and fungal (RE) Shannon and Simpson indexes of C. caput-medusae were significantly lower as compared to those of A. sparsifolia and T. ramosissima. Additionally, bacterial and fungal (RE and RS) node numbers and average degree of C. caput-medusae were lower than those found in A. sparsifolia and T. ramosissima. Root and soil nutrients collectively contributed to the composition of root-associated bacterial (RE, 12.4%; RS, 10.6%; BS, 16.6%) and fungal communities (RE, 34.3%; RS, 1.5%; BS, 17.7%). These findings demonstrate variations in the bacterial and fungal populations across different plant species with distinct compartments (RE, RS, and BS) in arid environments. More importantly, the study highlights how much soil and plant nutrients contribute to root-associated microbial communities.
Keywords: arid environments; desert plants; microbial communities; rhizosphere soil; soil nutrients.
Conflict of interest statement
There are no competing interests among the authors.
Figures




References
-
- Pankaj T., Jan E., Leach S.G.T., Tongmin S., Brajesh K.S. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020;18:607–621. - PubMed
-
- Gukailin A., Qin W.K., Wang X.D., Yu M., Feng J.G., Han M.G., Zhu B. Linking the rhizosphere effects of 12 woody species on soil microbial activities with soil and root nitrogen status. Rhizosphere. 2023;28:100809
-
- Mueller C.W., Baumert V., Carminati A., Germon A., Holz M., Kögel-Knabner I., Peth S., Schlüter S., Uteau D., Vetterlein D., et al. From rhizosphere to detritusphere-Soil structure formation driven by plant roots and the interactions with soil biota. Soil Biol. Biochem. 2024;193:109396. doi: 10.1016/j.soilbio.2024.109396. - DOI
LinkOut - more resources
Full Text Sources

