In Silico Molecular Modeling of Four New Afatinib Derived Molecules Targeting the Inhibition of the Mutated Form of BCR-ABL T315I
- PMID: 39275102
- PMCID: PMC11397288
- DOI: 10.3390/molecules29174254
In Silico Molecular Modeling of Four New Afatinib Derived Molecules Targeting the Inhibition of the Mutated Form of BCR-ABL T315I
Abstract
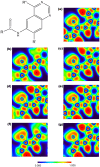
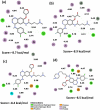
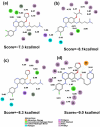
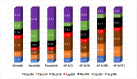
Four afatinib derivatives were designed and modeled. These derivatives were compared to the known tyrosine-kinase inhibitors in treating Chronic Myeloid Leukemia, i.e., imatinib and ponatinib. The molecules were evaluated through computational methods, including docking studies, the non-covalent interaction index, Electron Localization and Fukui Functions, in silico ADMET analysis, QTAIM, and Heat Map analysis. The AFA(IV) candidate significantly increases the score value compared to afatinib. Furthermore, AFA(IV) was shown to be relatively similar to the ponatinib profile when evaluating a range of molecular descriptors. The addition of a methylpiperazine ring seems to be well distributed in the structure of afatinib when targeting the BCR-ABL enzyme, providing an important hydrogen bond interaction with the Asp381 residue of the DFG-switch of BCR-ABL active site residue and the AFA(IV) new chemical entities. Finally, in silico toxicity predictions show a favorable index, with some molecules presenting the loss of the irritant properties associated with afatinib in theoretical predictions.
Keywords: BCR-ABL; CML; NCI; TKI; molecular modeling.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

