Peptides inhibiting the assembly of monomeric human l-lactate dehydrogenase into catalytically active homotetramer decrease the synthesis of lactate in cultured cells
- PMID: 39276013
- PMCID: PMC11400633
- DOI: 10.1002/pro.5161
Peptides inhibiting the assembly of monomeric human l-lactate dehydrogenase into catalytically active homotetramer decrease the synthesis of lactate in cultured cells
Abstract
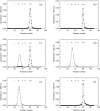
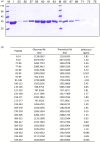
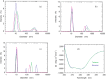
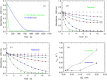
The energetic metabolism of cancer cells relies on a substantial commitment of pyruvate to the catalytic action of lactate-generating dehydrogenases. This coupling mainly depends on lactate dehydrogenase A (LDH-A), which is overexpressed in different types of cancers, and therefore represents an appealing therapeutic target. Taking into account that the activity of LDHs is exclusively exerted by their tetrameric forms, it was recently shown that peptides perturbing the monomers-to-tetramer assembly inhibit human LDH-A (hLDH-A). However, to identify these peptides, tetrameric hLDH-A was transiently exposed to strongly acidic conditions inducing its dissociation into monomers, which were tested as a target for peptides at low pH. Nevertheless, the availability of native monomeric hLDH-A would allow performing similar screenings under physiological conditions. Here we report on the unprecedented isolation of recombinant monomeric hLDH-A at neutral pH, and on its use to identify peptides inhibiting the assembly of the tetrameric enzyme. Remarkably, the GQNGISDL octapeptide, mimicking the 296-303 portion of hLDH-A C-terminal region, was observed to effectively inhibit the target enzyme. Moreover, by dissecting the action of this octapeptide, the cGQND cyclic tetrapeptide was found to act as the parental compound. Furthermore, we performed assays using MCF7 and BxPC3 cultured cells, exclusively expressing hLDH-A and hLDH-B, respectively. By means of these assays we detected a selective action of linear and cyclic GQND tetrapeptides, inhibiting lactate secretion in MCF7 cells only. Overall, our observations suggest that peptides mimicking the C-terminal region of hLDH-A effectively interfere with protein-protein interactions responsible for the assembly of the tetrameric enzyme.
Keywords: human lactate dehydrogenase A; inhibition of assembly; monomer; peptides; subunit–subunit interaction.
© 2024 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Arai K, Ishimitsu T, Fushinobu S, Uchikoba H, Matsuzawa H, Taguchi H. Active and inactive state structures of unliganded Lactobacillus casei allosteric l‐lactate dehydrogenase. Proteins. 2009;78:681–694. - PubMed
-
- Bernofsky C, Wanda SC. Formation of reduced nicotinamide adenine dinucleotide peroxide. J Biol Chem. 1982;257:6809–6817. - PubMed
-
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal Biochem. 1976;72:248–254. - PubMed
-
- Brooks GA. Lactate: glycolytic end product and oxidative substrate during sustained exercise in mammals: the lactate shuttle. In: Gilles R, editor. Circulation, respiration, and metabolism: current comparative approaches. Verlag, Berlin: Springer; 1985. p. 208–218.
-
- Chan WWC, Mosbach K. Effects of subunit interactions on the activity of lactate dehydrogenase studied in immobilized enzyme systems. Biochemistry. 1976;15:4215–4222. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

