A gain-of-function mutation at the C-terminus of FT-D1 promotes heading by interacting with 14-3-3A and FDL6 in wheat
- PMID: 39276323
- PMCID: PMC11672752
- DOI: 10.1111/pbi.14474
A gain-of-function mutation at the C-terminus of FT-D1 promotes heading by interacting with 14-3-3A and FDL6 in wheat
Abstract
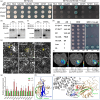
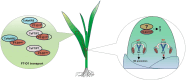
Vernalization and photoperiod pathways converging at FT1 control the transition to flowering in wheat. Here, we identified a gain-of-function mutation in FT-D1 that results in earlier heading date (HD), and shorter plant height and spike length in the gamma ray-induced eh1 wheat mutant. Knockout of the wild-type and overexpression of the mutated FT-D1 indicate that both alleles are functional to affect HD and plant height. Protein interaction assays demonstrated that the frameshift mutation in FT-D1eh1 exon 3 led to gain-of-function interactions with 14-3-3A and FDL6, thereby enabling the formation of florigen activation complex (FAC) and consequently activating a flowering-related transcriptomic programme. This mutation did not affect FT-D1eh1 interactions with TaNaKR5 or TaFTIP7, both of which could modulate HD, potentially via mediating FT-D1 translocation to the shoot apical meristem. Furthermore, the 'Segment B' external loop is essential for FT-D1 interaction with FDL6, while residue Y85 is required for interactions with TaNaKR5 and TaFTIP7. Finally, the flowering regulatory hub gene, ELF5, was identified as the FT-D1 regulatory target. This study illustrates FT-D1 function in determining wheat HD with a suite of interaction partners and provides genetic resources for tuning HD in elite wheat lines.
Keywords: ELF5; FAC; FT‐D1; heading date; plant height; wheat.
© 2024 The Author(s). Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Abe, M. , Kobayashi, Y. , Yamamoto, S. , Daimon, Y. , Yamaguchi, A. , Ikeda, Y. , Ichinoki, H. et al. (2005) FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309, 1052–1062. - PubMed
-
- Beales, J. , Turner, A. , GriYths, S. , Snape, J.W. and Laurie, D.A. (2007) A Pseudo‐Response Regulator is misexpressed in the photoperiod insensitive Ppd‐D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 115, 721–733. - PubMed
-
- Bonnin, I. , Rousset, M. , Madur, D. , Sourdille, P. , Dupuits, C. , Brunel, D. and Goldringer, I. (2007) FT genome A and D polymorphisms are associated with the variation of earliness components in hexaploid wheat. Theor. Appl. Genet. 116, 383–394. - PubMed
-
- Chen, X. , Xiong, H. , Guo, H. , Chen, S. , Zhao, L. , Xie, Y. , Gu, J. et al. (2024) Mapping and identification of a reverse mutation of Rht2 that enhances plant height and thousand grain weight in an elite wheat mutant induced by spaceflight. Plant Physiol. Biochem. 207, 108425. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

