Ligand-induced segregation from large cell-surface phosphatases is a critical step in γδ TCR triggering
- PMID: 39276348
- PMCID: PMC11452322
- DOI: 10.1016/j.celrep.2024.114761
Ligand-induced segregation from large cell-surface phosphatases is a critical step in γδ TCR triggering
Erratum in
-
Ligand-induced segregation from large cell-surface phosphatases is a critical step in γδ TCR triggering.Cell Rep. 2024 Oct 22;43(10):114882. doi: 10.1016/j.celrep.2024.114882. Epub 2024 Oct 8. Cell Rep. 2024. PMID: 39383038 No abstract available.
Abstract
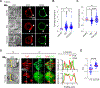
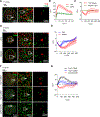
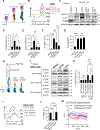
Gamma/delta (γδ) T cells are unconventional lymphocytes that recognize diverse ligands via somatically recombined T cell antigen receptors (γδ TCRs). The molecular mechanism by which ligand recognition initiates γδ TCR signaling, a process known as TCR triggering, remains elusive. Unlike αβ TCRs, γδ TCRs are not mechanosensitive and do not require co-receptors or typical binding-induced conformational changes for triggering. Here, we show that γδ TCR triggering by nonclassical MHC class Ib antigens, a major class of ligands recognized by γδ T cells, requires steric segregation of the large cell-surface phosphatases CD45 and CD148 from engaged TCRs at synaptic close-contact zones. Increasing access of these inhibitory phosphatases to sites of TCR engagement, by elongating MHC class Ib ligands or truncating CD45/148 ectodomains, abrogates TCR triggering and T cell activation. Our results identify a critical step in γδ TCR triggering and provide insight into the core triggering mechanism of endogenous and synthetic tyrosine-phosphorylated immunoreceptors.
Keywords: CD1d; CP: Immunology; T cell receptor; T22; nonclassical MHC antigens; triggering; γδ T cell.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests E.J.A. declares consultancy with Laguna Therapeutics, TcBioPharm, and Notch on γδ T cell immunotherapy development.
Figures




Update of
-
Ligand-induced segregation from large cell-surface phosphatases is a critical step in γδ TCR triggering.bioRxiv [Preprint]. 2023 Aug 24:2023.08.23.554524. doi: 10.1101/2023.08.23.554524. bioRxiv. 2023. Update in: Cell Rep. 2024 Sep 24;43(9):114761. doi: 10.1016/j.celrep.2024.114761. PMID: 37662246 Free PMC article. Updated. Preprint.
References
-
- Junqueira C, Polidoro RB, Castro G, Absalon S, Liang Z, Sen Santara S, Crespo A, Pereira DB, Gazzinelli RT, Dvorin JD, and Lieberman J (2021). gammadelta T cells suppress Plasmodium falciparum blood-stage infection by direct killing and phagocytosis. Nat Immunol 22, 347–357. 10.1038/s41590-020-00847-4. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

