Co-opting templated aggregation to degrade pathogenic tau assemblies and improve motor function
- PMID: 39276772
- PMCID: PMC7616835
- DOI: 10.1016/j.cell.2024.08.024
Co-opting templated aggregation to degrade pathogenic tau assemblies and improve motor function
Abstract
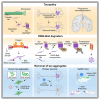
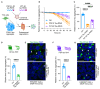
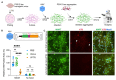
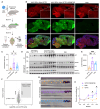
Protein aggregation causes a wide range of neurodegenerative diseases. Targeting and removing aggregates, but not the functional protein, is a considerable therapeutic challenge. Here, we describe a therapeutic strategy called "RING-Bait," which employs an aggregating protein sequence combined with an E3 ubiquitin ligase. RING-Bait is recruited into aggregates, whereupon clustering dimerizes the RING domain and activates its E3 function, resulting in the degradation of the aggregate complex. We exemplify this concept by demonstrating the specific degradation of tau aggregates while sparing soluble tau. Unlike immunotherapy, RING-Bait is effective against both seeded and cell-autonomous aggregation. RING-Bait removed tau aggregates seeded from Alzheimer's disease (AD) and progressive supranuclear palsy (PSP) brain extracts and was also effective in primary neurons. We used a brain-penetrant adeno-associated virus (AAV) to treat P301S tau transgenic mice, reducing tau pathology and improving motor function. A RING-Bait strategy could be applied to other neurodegenerative proteinopathies by replacing the Bait sequence to match the target aggregate.
Keywords: Alzheimer’s disease; TRIM21; antibodies; gene therapy; neurobiology; neurodegeneration; protein engineering; targeted protein degradation; tauopathy; ubiquitination.
Crown Copyright © 2024. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests L.V.C.M., G.P., W.A.M., and L.C.J. are listed as inventors on a patent containing data published in this paper.
Figures


References
-
- Goedert M. NEURODEGENERATION. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science. 2015;349:1255555. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

