Genome-wide discovery and integrative genomic characterization of insulin resistance loci using serum triglycerides to HDL-cholesterol ratio as a proxy
- PMID: 39277575
- PMCID: PMC11401929
- DOI: 10.1038/s41467-024-52105-y
Genome-wide discovery and integrative genomic characterization of insulin resistance loci using serum triglycerides to HDL-cholesterol ratio as a proxy
Abstract
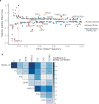
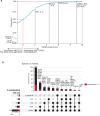
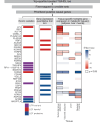
Insulin resistance causes multiple epidemic metabolic diseases, including type 2 diabetes, cardiovascular disease, and fatty liver, but is not routinely measured in epidemiological studies. To discover novel insulin resistance genes in the general population, we conducted genome-wide association studies in 382,129 individuals for triglyceride to HDL-cholesterol ratio (TG/HDL), a surrogate marker of insulin resistance calculable from commonly measured serum lipid profiles. We identified 251 independent loci, of which 62 were more strongly associated with TG/HDL compared to TG or HDL alone, suggesting them as insulin resistance loci. Candidate causal genes at these loci were prioritized by fine mapping with directions-of-effect and tissue specificity annotated through analysis of protein coding and expression quantitative trait variation. Directions-of-effect were corroborated in an independent cohort of individuals with directly measured insulin resistance. We highlight two phospholipase encoding genes, PLA2G12A and PLA2G6, which liberate arachidonic acid and improve insulin sensitivity, and VGLL3, a transcriptional co-factor that increases insulin resistance partially through enhanced adiposity. Finally, we implicate the anti-apoptotic gene TNFAIP8 as a sex-dimorphic insulin resistance factor, which acts by increasing visceral adiposity, specifically in females. In summary, our study identifies several candidate modulators of insulin resistance that have the potential to serve as biomarkers and pharmacological targets.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
P.N. reports research grants from Allelica, Apple, Amgen, Boston Scientific, Genentech/Roche, and Novartis, personal fees from Allelica, Apple, AstraZeneca, Blackstone Life Sciences, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co, Foresite Labs, Genentech/Roche, GV, HeartFlow, Magnet Biomedicine, and Novartis, scientific advisory board membership of Esperion Therapeutics, Preciseli, and TenSixteen Bio, scientific co-founder of TenSixteen Bio, equity in MyOme, Preciseli, and TenSixteen Bio, and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. The remaining authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

