The piranha gut microbiome provides a selective lens into river water biodiversity
- PMID: 39277613
- PMCID: PMC11401890
- DOI: 10.1038/s41598-024-72329-8
The piranha gut microbiome provides a selective lens into river water biodiversity
Abstract
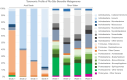
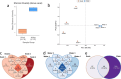
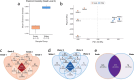
Advances in omics technologies have enabled the in-depth study of microbial communities and their metabolic profiles from all environments. Here metagenomes were sampled from piranha (Serrasalmus rhombeus) and from river water from the Rio São Benedito (Amazon Basin). Shotgun metagenome sequencing was used to explore diversity and to test whether fish microbiomes are a good proxy for river microbiome studies. The results showed that the fish microbiomes were not significantly different from the river water microbiomes at higher taxonomic ranks. However, at the genus level, fish microbiome alpha diversity decreased, and beta diversity increased. This result repeated for functional gene abundances associated with specific metabolic categories (SEED level 3). A clear delineation between water and fish was seen for beta diversity. The piranha microbiome provides a good and representative subset of its river water microbiome. Variations seen in beta biodiversity were expected and can be explained by temporal variations in the fish microbiome in response to stronger selective forces on its biodiversity. Metagenome assembled genomes construction was better from the fish samples. This study has revealed that the microbiome of a piranha tells us a lot about its river water microbiome and function.
Keywords: Serrasalmus rhombeus; Amazon river microbiome; Biotechnology; Host-associated microbiome; Metagenomics.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Towards a unified medical microbiome ecology of the OMU for metagenomes and the OTU for microbes.BMC Bioinformatics. 2024 Mar 29;25(1):137. doi: 10.1186/s12859-023-05591-8. BMC Bioinformatics. 2024. PMID: 38553666 Free PMC article.
-
Microbiome Analysis Reveals Diversity and Function of Mollicutes Associated with the Eastern Oyster, Crassostrea virginica.mSphere. 2021 May 12;6(3):e00227-21. doi: 10.1128/mSphere.00227-21. mSphere. 2021. PMID: 33980678 Free PMC article.
-
Strain Structure and Dynamics Revealed by Targeted Deep Sequencing of the Honey Bee Gut Microbiome.mSphere. 2020 Aug 26;5(4):e00694-20. doi: 10.1128/mSphere.00694-20. mSphere. 2020. PMID: 32848005 Free PMC article.
-
Genome-resolved metagenomics: a game changer for microbiome medicine.Exp Mol Med. 2024 Jul;56(7):1501-1512. doi: 10.1038/s12276-024-01262-7. Epub 2024 Jul 1. Exp Mol Med. 2024. PMID: 38945961 Free PMC article. Review.
-
Omics in gut microbiome analysis.J Microbiol. 2021 Mar;59(3):292-297. doi: 10.1007/s12275-021-1004-0. Epub 2021 Feb 23. J Microbiol. 2021. PMID: 33624266 Review.
References
-
- Wani, A. K. et al. Bioprospecting culturable and unculturable microbial consortia through metagenomics for bioremediation. Clean. Chem. Eng.2, 100017 (2022).10.1016/j.clce.2022.100017 - DOI
MeSH terms
Grants and funding
- Bolsa de Doutorado/Conselho Nacional de Desenvolvimento Científico e Tecnológico
- Bolsa de Iniciacao Cientifica/Conselho Nacional de Desenvolvimento Científico e Tecnológico
- Bolsa de doutordo/Coordenação de Aperfeiçoamento de Pessoal de Nível Superior
- PRINT 001/Coordenação de Aperfeiçoamento de Pessoal de Nível Superior
LinkOut - more resources
Full Text Sources

