The adult shell matrix protein repertoire of the marine snail Crepidula is dominated by conserved genes that are also expressed in larvae
- PMID: 39277725
- PMCID: PMC11401363
- DOI: 10.1186/s12862-024-02237-y
The adult shell matrix protein repertoire of the marine snail Crepidula is dominated by conserved genes that are also expressed in larvae
Abstract
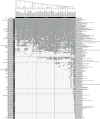
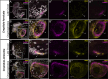
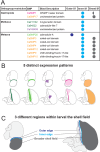
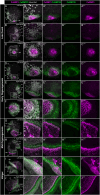
Mollusca is a morphologically diverse phylum, exhibiting an immense variety of calcium carbonate structures. Proteomic studies of adult shells often report high levels of rapidly-evolving, 'novel' shell matrix proteins (SMPs), which are hypothesized to drive shell diversification. However, relatively little is known about the phylogenetic distribution of SMPs, or about the function of individual SMPs in shell construction. To understand how SMPs contribute to shell diversification a thorough characterization of SMPs is required. Here, we build tools and a foundational understanding of SMPs in the marine gastropod species Crepidula fornicata and Crepidula atrasolea because they are genetically-enabled mollusc model organisms. First, we established a staging system of shell development in C. atrasolea for the first time. Next, we leveraged previous findings in C. fornicata combined with phylogenomic analyses of 95 metazoan species to determine the evolutionary lineage of its adult SMP repertoire. We found that 55% of C. fornicata's SMPs belong to molluscan orthogroups, with 27% restricted to Gastropoda, and only 5% restricted at the species level. The low percentage of species-restricted SMPs underscores the importance of broad-taxon sampling and orthology inference approaches when determining homology of SMPs. From our transcriptome analysis, we found that the majority of C. fornicata SMPs that were found conserved in C. atrasolea were expressed in both larval and adult stages. We then selected a subset of SMPs of varying evolutionary ages for spatial-temporal analysis using in situ hybridization chain reaction (HCR) during larval shell development in C. atrasolea. Out of the 18 SMPs analyzed, 12 were detected in the larval shell field. These results suggest overlapping larval vs. adult SMP repertoires. Using multiplexed HCR, we observed five SMP expression patterns and three distinct cell populations within the shell field. These patterns support the idea that modular expression of SMPs could facilitate divergence of shell morphological characteristics. Collectively, these data establish an evolutionary and developmental framework in Crepidula that enables future comparisons of molluscan biomineralization to reveal mechanisms of shell diversification.
Keywords: Biomineralization; Crepidula; Shell development; Shell matrix proteins.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
CRISPR/Cas9 Knockout of Shell Matrix Protein 1 in the Slipper-Snail Crepidula atrasolea.J Exp Zool B Mol Dev Evol. 2025 Jul;344(5):266-283. doi: 10.1002/jez.b.23293. Epub 2025 May 4. J Exp Zool B Mol Dev Evol. 2025. PMID: 40320697 Free PMC article.
-
Spatial-temporal expression analysis of lineage-restricted shell matrix proteins reveals shell field regionalization and distinct cell populations in the slipper snail Crepidula atrasolea.bioRxiv [Preprint]. 2023 Mar 21:2023.03.18.532128. doi: 10.1101/2023.03.18.532128. bioRxiv. 2023. PMID: 36993573 Free PMC article. Preprint.
-
Proteomic and Transcriptomic Analyses in the Slipper Snail Crepidula fornicata Uncover Shell Matrix Genes Expressed During Adult and Larval Biomineralization.Integr Org Biol. 2022 Aug 10;4(1):obac023. doi: 10.1093/iob/obac023. eCollection 2022. Integr Org Biol. 2022. PMID: 35968217 Free PMC article.
-
Review: Post-translational modifications of marine shell matrix proteins.Comp Biochem Physiol B Biochem Mol Biol. 2021 Oct-Dec;256:110641. doi: 10.1016/j.cbpb.2021.110641. Epub 2021 Jun 25. Comp Biochem Physiol B Biochem Mol Biol. 2021. PMID: 34182126 Review.
-
Molluscan models: Crepidula fornicata.Curr Opin Genet Dev. 2016 Aug;39:138-148. doi: 10.1016/j.gde.2016.05.021. Epub 2016 Aug 12. Curr Opin Genet Dev. 2016. PMID: 27526387 Review.
Cited by
-
CRISPR/Cas9 Knockout of Shell Matrix Protein 1 in the Slipper-Snail Crepidula atrasolea.J Exp Zool B Mol Dev Evol. 2025 Jul;344(5):266-283. doi: 10.1002/jez.b.23293. Epub 2025 May 4. J Exp Zool B Mol Dev Evol. 2025. PMID: 40320697 Free PMC article.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
