Interpretable discriminant analysis for functional data supported on random nonlinear domains with an application to Alzheimer's disease
- PMID: 39279915
- PMCID: PMC11398888
- DOI: 10.1093/jrsssb/qkae023
Interpretable discriminant analysis for functional data supported on random nonlinear domains with an application to Alzheimer's disease
Abstract
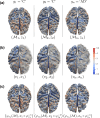
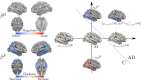
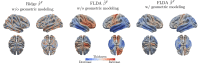
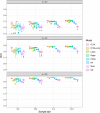
We introduce a novel framework for the classification of functional data supported on nonlinear, and possibly random, manifold domains. The motivating application is the identification of subjects with Alzheimer's disease from their cortical surface geometry and associated cortical thickness map. The proposed model is based upon a reformulation of the classification problem as a regularized multivariate functional linear regression model. This allows us to adopt a direct approach to the estimation of the most discriminant direction while controlling for its complexity with appropriate differential regularization. Our approach does not require prior estimation of the covariance structure of the functional predictors, which is computationally prohibitive in our application setting. We provide a theoretical analysis of the out-of-sample prediction error of the proposed model and explore the finite sample performance in a simulation setting. We apply the proposed method to a pooled dataset from Alzheimer's Disease Neuroimaging Initiative and Parkinson's Progression Markers Initiative. Through this application, we identify discriminant directions that capture both cortical geometric and thickness predictive features of Alzheimer's disease that are consistent with the existing neuroscience literature.
Keywords: functional classification; manifold data analysis; neuroimaging; shape data analysis.
© The Royal Statistical Society 2024.
Conflict of interest statement
Conflict of interest: None declared.
Figures


References
-
- Berlinet A., & Thomas-Agnan C (2004). Reproducing kernel Hilbert spaces in probability and statistics. Springer US. 10.1007/978-1-4419-9096-9 - DOI
-
- Berrendero J. R., Cuevas A., & Torrecilla J. L. (2018). On the use of reproducing kernel Hilbert spaces in functional classification. Journal of the American Statistical Association, 113(523), 1210–1218. 10.1080/01621459.2017.1320287 - DOI
-
- Biffi C., De Marvao A., Attard M. I., Dawes T. J., Whiffin N., Bai W., Shi W., Francis C., Meyer H., Buchan R., Cook S. A., Rueckert D., & O’Regan D. P. (2018). Three-dimensional cardiovascular imaging-genetics: A mass univariate framework. Bioinformatics, 34(1), 97–103. 10.1093/bioinformatics/btx552 - DOI - PMC - PubMed
-
- Blanchard, G., & Krämer, N. (2010). Optimal learning rates for kernel conjugate gradient regression. In Proceedings of the 23rd international conference on neural information processing systems – volume 1 (NIPS'10) (pp. 226–234). Curran Associates Inc., Red Hook, NY, USA. 10.5555/2997189.2997215 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
