Liquid-liquid phase separation-related features of PYGB/ACTR3/CCNA2/ITGB1/ATP8A1/RAP1GAP2 predict the prognosis of pancreatic cancer
- PMID: 39279964
- PMCID: PMC11399862
- DOI: 10.21037/jgo-24-426
Liquid-liquid phase separation-related features of PYGB/ACTR3/CCNA2/ITGB1/ATP8A1/RAP1GAP2 predict the prognosis of pancreatic cancer
Abstract
Background: The growth and metastasis of pancreatic cancer (PC) has been found to be closely associated with liquid-liquid phase separation (LLPS). This study sought to identify LLPS-related biomarkers in PC to construct a robust prognostic model.
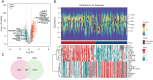
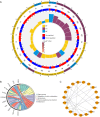
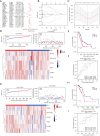
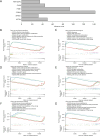
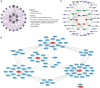
Methods: Transcriptomic data and clinical information related to PC were retrieved from publicly accessible databases. The PC-related data set was subjected to differential expression, Mendelian randomization (MR), univariate Cox, and least absolute selection and shrinkage operator analyses to identify biomarkers. Using the biomarkers, we subsequently constructed a risk model, identified the independent prognostic factors of PC, established a nomogram, and conducted an immune analysis.
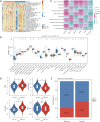
Results: The study identified four genes linked with an increased risk of PC; that is, PYGB, ACTR3, CCNA2, and ITGB1. Conversely, ATP8A1, and RAP1GAP2 were found to provide protection against PC. These findings contributed significantly to the development of a highly precise risk model in which risk, age, and pathology N stage were categorized as independent factors in predicting the prognosis of PC patients. Using these factors, a nomogram was established to predict survival outcomes accurately. An immune analysis revealed varying levels of eosinophils, gamma delta T cells, and other immune cells between the distinct risk groups. The high-risk patients exhibited increased potential for immune escape, while the low-risk patients showed a higher response to immunotherapy.
Conclusions: Six genes were identified as having potential causal relationships with PC. These genes were integrated into a prognostic risk model, thereby serving as unique prognostic signatures. Our findings provide novel insights into predicting the prognosis of PC patients.
Keywords: Mendelian randomization (MR); Pancreatic cancer (PC); biomarkers; liquid-liquid phase separation (LLPS); prognosis.
2024 Journal of Gastrointestinal Oncology. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-24-426/coif). The authors have no conflicts of interest to declare.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous
