5-Fluorouracil treatment represses pseudouridine-containing miRNA export into extracellular vesicles
- PMID: 39281020
- PMCID: PMC11393769
- DOI: 10.1002/jex2.70010
5-Fluorouracil treatment represses pseudouridine-containing miRNA export into extracellular vesicles
Abstract
5-Fluorouracil (5-FU) has been used for chemotherapy for colorectal and other cancers for over 50 years. The prevailing view of its mechanism of action is inhibition of thymidine synthase leading to defects in DNA replication and repair. However, 5-FU is also incorporated into RNA causing defects in RNA metabolism, inhibition of pseudouridine modification, and altered ribosome function. We examined the impact of 5-FU on post-transcriptional small RNA modifications (PTxMs) and the expression and export of RNA into small extracellular vesicles (sEVs). EVs are secreted by all cells and contain a variety of proteins and RNAs that can function in cell-cell communication. We found that treatment of colorectal cancer (CRC) cells with 5-FU represses sEV export of miRNA and snRNA-derived RNAs, but promotes export of snoRNA-derived RNAs. Strikingly, 5-FU treatment significantly decreased the levels of pseudouridine on both cellular and sEV small RNA profiles. In contrast, 5-FU exposure led to increased levels of cellular small RNAs containing a variety of methyl-modified bases. These unexpected findings show that 5-FU exposure leads to altered RNA expression, base modification, and aberrant trafficking and localization of small RNAs.
Keywords: 5‐FU; EV export; RNA modification; extracellular vesicles; miRNA; pseudourine.
© 2024 The Author(s). Journal of Extracellular Biology published by Wiley Periodicals LLC on behalf of International Society for Extracellular Vesicles.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
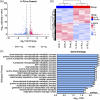


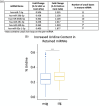

Update of
-
5-Fluorouracil Treatment Represses Pseudouridine-Containing Small RNA Export into Extracellular Vesicles.bioRxiv [Preprint]. 2024 Jan 16:2024.01.15.575751. doi: 10.1101/2024.01.15.575751. bioRxiv. 2024. Update in: J Extracell Biol. 2024 Sep 13;3(9):e70010. doi: 10.1002/jex2.70010. PMID: 38293013 Free PMC article. Updated. Preprint.
References
-
- Abe, Y. , Sakuyama, N. , Sato, T. , Kishine, K. , Nagayasu, K. , Nakatani, A. , Kitajima, M. , Watanabe, T. , Nishimura, K. , Ochiai, T. , & Nagaoka, I. (2019). Evaluation of the 5‐fluorouracil plasma level in patients with colorectal cancer undergoing continuous infusion chemotherapy. Molecular and Clinical Oncology, 11, 289–295. - PMC - PubMed
-
- Abner, J. J. , Franklin, J. L. , Clement, M. A. , Hinger, S. A. , Allen, R. M. , Liu, X. , Kellner, S. , Wu, J. , Karijolich, J. , Liu, Q. , Vickers, K. C. , Dedon, P. , Weaver, A. M. , Coffey, R. J. , & Patton, J. G. (2021). Depletion of METTL3 alters cellular and extracellular levels of miRNAs containing m(6)A consensus sequences. Heliyon, 7, e08519. - PMC - PubMed
-
- Allen, R. M. , Michell, D. L. , Cavnar, A. B. , Zhu, W. , Makhijani, N. , Contreras, D. M. , Raby, C. A. , Semler, E. M. , DeJulius, C. , Castleberry, M. , Zhang, Y. , Ramirez‐Solano, M. , Zhao, S. , Duvall, C. , Doran, A. C. , Sheng, Q. , Linton, M. F. , & Vickers, K. C. (2022). LDL delivery of microbial small RNAs drives atherosclerosis through macrophage TLR8. Nature Cell Biology, 24, 1701–1713. - PMC - PubMed
-
- Allen, R. M. , Zhao, S. , Ramirez Solano, M. A. , Zhu, W. , Michell, D. L. , Wang, Y. , Shyr, Y. , Sethupathy, P. , Linton, M. F. , Graf, G. A. , Sheng, Q. , & Vickers, K. C. (2018). Bioinformatic analysis of endogenous and exogenous small RNAs on lipoproteins. Journal of Extracellular Vesicles, 7, 1506198. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
