Rigid crosslinking of the CD3 complex leads to superior T cell stimulation
- PMID: 39281668
- PMCID: PMC11392757
- DOI: 10.3389/fimmu.2024.1434463
Rigid crosslinking of the CD3 complex leads to superior T cell stimulation
Abstract
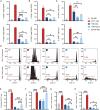
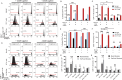
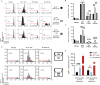
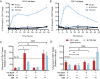
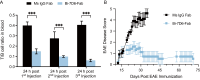
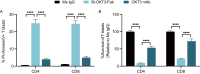
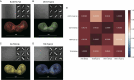
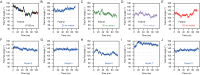
Functionally bivalent non-covalent Fab dimers (Bi-Fabs) specific for the TCR/CD3 complex promote CD3 signaling on T cells. While comparing functional responses to stimulation with Bi-Fab, F(ab')2 or mAb specific for the same CD3 epitope, we observed fratricide requiring anti-CD3 bridging of adjacent T cells. Surprisingly, anti-CD3 Bi-Fab ranked first in fratricide potency, followed by anti-CD3 F(ab')2 and anti-CD3 mAb. Low resolution structural studies revealed anti-CD3 Bi-Fabs and F(ab')2 adopt similar global shapes with CD3-binding sites oriented outward. However, under molecular dynamic simulations, anti-CD3 Bi-Fabs crosslinked CD3 more rigidly than F(ab')2. Furthermore, molecular modelling of Bi-Fab and F(ab')2 binding to CD3 predicted crosslinking of T cell antigen receptors located in opposing plasma membrane domains, a feature fitting with T cell fratricide observed. Thus, increasing rigidity of Fab-CD3 crosslinking between opposing effector-target pairs may result in stronger T cell effector function. These findings could guide improving clinical performance of bi-specific anti-CD3 drugs.
Keywords: CD3/antibody crosslinking; EAE (experimental autoimmune encephalomyelitis); T cell division and apoptosis; T cell receptor engagement and triggering; anti-CD3 Fab-based therapies; antibody fragment structure; molecular dynamic simulation.
Copyright © 2024 Nelson, Wang, Laffey, Becher, Parks, Hoffmann, Galeano, Mangalam, Teixeiro, White, Schrum, Cannon and Gil.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

