The influence of 4G/5G polymorphism in the plasminogen-activator-inhibitor-1 promoter on COVID-19 severity and endothelial dysfunction
- PMID: 39281671
- PMCID: PMC11392769
- DOI: 10.3389/fimmu.2024.1445294
The influence of 4G/5G polymorphism in the plasminogen-activator-inhibitor-1 promoter on COVID-19 severity and endothelial dysfunction
Abstract
Introduction: Plasminogen activator inhibitor-1 (PAI-1) is linked to thrombosis and endothelial dysfunction in severe COVID-19. The +43 G>A PAI-1 and 4G/5G promoter polymorphism can influence PAI-1 expression. The 4G5G PAI-1 promoter gene polymorphism constitutes the 4G4G, 4G5G, and 5G5G genotypes. However, the impact of PAI-1 polymorphisms on disease severity or endothelial dysfunction remains unclear.
Methods: Clinical data, sera, and peripheral blood mononuclear cells (PBMCs) of COVID-19 patients were studied.
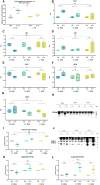
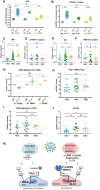
Results: Comorbidities and clinical biomarkers did not correlate with genotypes in either polymorphism. However, differences between fibrinolytic factors and interleukin-1β (IL-1β) were identified in genotypes of the 4G/5G but not the 43 G>A PAI polymorphism. Patients with the 4G4G genotype of the 4G/5G polymorphism showed high circulating PAI-1, mainly complexed with plasminogen activators, and low IL-1β and plasmin levels, indicating suppressed fibrinolysis. NFκB was upregulated in PBMCs of COVID-19 patients with the 4G4G genotype.
Discussion: Mechanistically, IL-1β enhanced PAI-1 expression in 4G4G endothelial cells, preventing the generation of plasmin and cleavage products like angiostatin, soluble uPAR, and VCAM1. We identified inflammation-induced endothelial dysfunction coupled with fibrinolytic system overactivation as a risk factor for patients with the 5G5G genotype.
Keywords: COVID-19; PAI-1 4G/5G promoter polymorphism (rs1799889); PAI-1 polymorphism +43G>A (rs6092); endothelial cells; inflammation; plasmin; interleukin-1-β; plasminogen activator inhibitor-1.
Copyright © 2024 Yatsenko, Rios, Nogueira, Salama, Takahashi, Adachi, Tabe, Hattori, Osada, Naito, Takahashi, Hattori and Heissig.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Keragala CB, Medcalf RL. Plasminogen: an enigmatic zymogen. Blood 137(21):2881–9. - PubMed
-
- Munakata S, Tashiro Y, Nishida C, Sato A, Komiyama H, Shimazu H, et al. Inhibition of plasmin protects against colitis in mice by suppressing matrix metalloproteinase 9-mediated cytokine release from myeloid cells. Gastroenterology. (2015) 148:565–78.e4. doi: 10.1053/j.gastro.2014.12.001 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

