Nanobody against SARS-CoV-2 non-structural protein Nsp9 inhibits viral replication in human airway epithelia
- PMID: 39281707
- PMCID: PMC11401216
- DOI: 10.1016/j.omtn.2024.102304
Nanobody against SARS-CoV-2 non-structural protein Nsp9 inhibits viral replication in human airway epithelia
Abstract
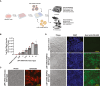
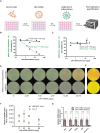
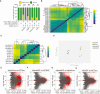
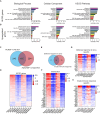
Nanobodies are emerging as critical tools for drug design. Several have been recently created to serve as inhibitors of severe acute respiratory syndrome coronavirus s (SARS-CoV-2) entry in the host cell by targeting surface-exposed spike protein. Here we have established a pipeline that instead targets highly conserved viral proteins made only after viral entry into the host cell when the SARS-CoV-2 RNA-based genome is translated. As proof of principle, we designed nanobodies against the SARS-CoV-2 non-structural protein (Nsp)9, which is required for viral genome replication. One of these anti-Nsp9 nanobodies, 2NSP23, previously characterized using immunoassays and nuclear magnetic resonance spectroscopy for epitope mapping, was expressed and found to block SARS-CoV-2 replication specifically. We next encapsulated 2NSP23 nanobody into lipid nanoparticles (LNPs) as mRNA. We show that this nanobody, hereby referred to as LNP-mRNA-2NSP23, is internalized and translated in cells and suppresses multiple SARS-CoV-2 variants, as seen by qPCR and RNA deep sequencing. These results are corroborated in three-dimensional reconstituted human epithelium kept at air-liquid interface to mimic the outer surface of lung tissue. These observations indicate that LNP-mRNA-2NSP23 is internalized and, after translation, it inhibits viral replication by targeting Nsp9 in living cells. We speculate that LNP-mRNA-2NSP23 may be translated into an innovative strategy to generate novel antiviral drugs highly efficient across coronaviruses.
Keywords: MT: LNP-based Delivery Strategies; Non-structural protein 9; Nsp9; SARS-CoV-2; innate immunity; nanobody.
© 2024 The Author(s).
Conflict of interest statement
P.P., G.E., H.T.H., and S.C.D. are part of a US patent filed by New York University in Abu Dhabi jointly with ISAR Biosciences.
Figures




References
-
- Woo P.C.Y., Lau S.K.P., Chu C.M., Chan K.H., Tsoi H.W., Huang Y., Wong B.H.L., Poon R.W.S., Cai J.J., Luk W.K., et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 2005;79:884–895. doi: 10.1128/JVI.79.2.884-895.2005. - DOI - PMC - PubMed
-
- Matoba Y., Abiko C., Ikeda T., Aoki Y., Suzuki Y., Yahagi K., Matsuzaki Y., Itagaki T., Katsushima F., Katsushima Y., Mizuta K. Detection of the human coronavirus 229E, HKU1, NL63, and OC43 between 2010 and 2013 in Yamagata, Japan. Jpn. J. Infect. Dis. 2015;68:138–141. doi: 10.7883/yoken.JJID.2014.266. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

