This is a preprint.
Analysis of more than 400,000 women provides case-control evidence for BRCA1 and BRCA2 variant classification
- PMID: 39281752
- PMCID: PMC11398439
- DOI: 10.1101/2024.09.04.24313051
Analysis of more than 400,000 women provides case-control evidence for BRCA1 and BRCA2 variant classification
Update in
-
Analysis of more than 400,000 women provides case-control evidence for BRCA1 and BRCA2 variant classification.Nat Commun. 2025 May 25;16(1):4852. doi: 10.1038/s41467-025-59979-6. Nat Commun. 2025. PMID: 40413188 Free PMC article.
Abstract
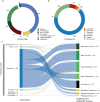
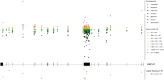
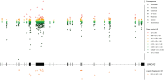
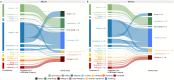
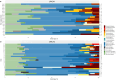
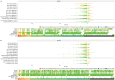
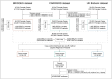
Clinical genetic testing identifies variants causal for hereditary cancer, information that is used for risk assessment and clinical management. Unfortunately, some variants identified are of uncertain clinical significance (VUS), complicating patient management. Case-control data is one evidence type used to classify VUS, and previous findings indicate that case-control likelihood ratios (LRs) outperform odds ratios for variant classification. As an initiative of the Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA) Analytical Working Group we analyzed germline sequencing data of BRCA1 and BRCA2 from 96,691 female breast cancer cases and 303,925 unaffected controls from three studies: the BRIDGES study of the Breast Cancer Association Consortium, the Cancer Risk Estimates Related to Susceptibility consortium, and the UK Biobank. We observed 11,227 BRCA1 and BRCA2 variants, with 6,921 being coding, covering 23.4% of BRCA1 and BRCA2 VUS in ClinVar and 19.2% of ClinVar curated (likely) benign or pathogenic variants. Case-control LR evidence was highly consistent with ClinVar assertions for (likely) benign or pathogenic variants; exhibiting 99.1% sensitivity and 95.4% specificity for BRCA1 and 92.2% sensitivity and 86.6% specificity for BRCA2. This approach provides case-control evidence for 785 unclassified variants, that can serve as a valuable element for clinical classification.
Keywords: BRCA1; BRCA2; VUS; case-control; likelihood ratio; odds ratio; rare variant.
Figures

References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
