This is a preprint.
Connecting genomic and proteomic signatures of amyloid burden in the brain
- PMID: 39281766
- PMCID: PMC11398581
- DOI: 10.1101/2024.09.06.24313124
Connecting genomic and proteomic signatures of amyloid burden in the brain
Abstract
Background: Alzheimer's disease (AD) has a high heritable component characteristic of complex diseases, yet many of the genetic risk factors remain unknown. We combined genome-wide association studies (GWAS) on amyloid endophenotypes measured in cerebrospinal fluid (CSF) and positron emission tomography (PET) as surrogates of amyloid pathology, which may be helpful to understand the underlying biology of the disease.
Methods: We performed a meta-analysis of GWAS of CSF Aβ42 and PET measures combining six independent cohorts (n=2,076). Due to the opposite effect direction of Aβ phenotypes in CSF and PET measures, only genetic signals in the opposite direction were considered for analysis (n=376,599). Polygenic risk scores (PRS) were calculated and evaluated for AD status and amyloid endophenotypes. We then searched the CSF proteome signature of brain amyloidosis using SOMAscan proteomic data (Ace cohort, n=1,008) and connected it with GWAS results of loci modulating amyloidosis. Finally, we compared our results with a large meta-analysis using publicly available datasets in CSF (n=13,409) and PET (n=13,116). This combined approach enabled the identification of overlapping genes and proteins associated with amyloid burden and the assessment of their biological significance using enrichment analyses.
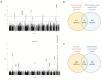
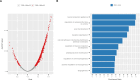
Results: After filtering the meta-GWAS, we observed genome-wide significance in the rs429358-APOE locus and nine suggestive hits were annotated. We replicated the APOE loci using the large CSF-PET meta-GWAS and identified multiple AD-associated genes as well as the novel GADL1 locus. Additionally, we found a significant association between the AD PRS and amyloid levels, whereas no significant association was found between any Aβ PRS with AD risk. CSF SOMAscan analysis identified 1,387 FDR-significant proteins associated with CSF Aβ42 levels. The overlap among GWAS loci and proteins associated with amyloid burden was very poor (n=35). The enrichment analysis of overlapping hits strongly suggested several signalling pathways connecting amyloidosis with the anchored component of the plasma membrane, synapse physiology and mental disorders that were replicated in the large CSF-PET meta-analysis.
Conclusions: The strategy of combining CSF and PET amyloid endophenotypes GWAS with CSF proteome analyses might be effective for identifying signals associated with the AD pathological process and elucidate causative molecular mechanisms behind the amyloid mobilization in AD.
Keywords: Aβ42; CSF biomarkers; GWAS; PET tomography; Proteome.
Conflict of interest statement
Competing interests All authors declare that the research was conducted in the absence of any conflict of interest.
Figures




References
-
- 2022 Alzheimer’s disease Facts and Figures: More Than Normal Aging: Understanding Mild Cognitive Impairment. Alzheimer’s & Dementia. 2022;18(4):700–789. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
