This is a preprint.
Genome-wide association studies of Down syndrome associated congenital heart defects
- PMID: 39281767
- PMCID: PMC11398599
- DOI: 10.1101/2024.09.06.24313183
Genome-wide association studies of Down syndrome associated congenital heart defects
Update in
-
Genome-Wide Association Studies of Down Syndrome Associated Congenital Heart Defects Suggests a Genetically Heterogeneous Risk for CHD in DS.Genet Epidemiol. 2025 Jun;49(4):e70010. doi: 10.1002/gepi.70010. Genet Epidemiol. 2025. PMID: 40407036 Free PMC article.
Abstract
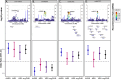
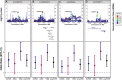
Congenital heart defects (CHDs) are the most common structural birth defect and are present in 40-50% of children born with Down syndrome (DS). To characterize the genetic architecture of DS-associated CHD, we sequenced genomes of a multiethnic group of children with DS and a CHD (n=886: atrioventricular septal defects (AVSD), n=438; atrial septal defects (ASD), n=122; ventricular septal defects (VSD), n=170; other types of CHD, n=156) and DS with a structurally normal heart (DS+NH, n=572). We performed four GWAS for common variants (MAF>0.05) comparing DS with CHD, stratified by CHD-subtype, to DS+NH controls. Although no SNP achieved genome-wide significance, multiple loci in each analysis achieved suggestive significance (p<2×10-6). Of these, the 1p35.1 locus (near RBBP4) was specifically associated with ASD risk and the 5q35.2 locus (near MSX2) was associated with any type of CHD. Each of the suggestive loci contained one or more plausible candidate genes expressed in the developing heart. While no SNP replicated (p<2×10-6) in an independent cohort of DS+CHD (DS+CHD: n=229; DS+NH: n=197), most SNPs that were suggestive in our GWASs remained suggestive when meta-analyzed with the GWASs from the replication cohort. These results build on previous work to identify genetic modifiers of DS-associated CHD.
Keywords: Down syndrome; birth defect; congenital heart defect; genome-wide association study; trisomy 21.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
