Auto-expansion of in vivo HDAd-transduced hematopoietic stem cells by constitutive expression of tHMGA2
- PMID: 39282078
- PMCID: PMC11399618
- DOI: 10.1016/j.omtm.2024.101319
Auto-expansion of in vivo HDAd-transduced hematopoietic stem cells by constitutive expression of tHMGA2
Abstract
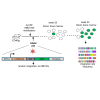
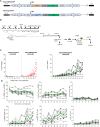
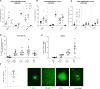
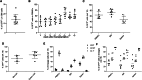
We developed an in vivo hematopoietic stem cell (HSC) gene therapy approach that does not require cell transplantation. To achieve therapeutically relevant numbers of corrected cells, we constructed HSC-tropic HDAd5/35++ vectors expressing a 3' UTR truncated HMGA2 gene and a GFP reporter gene. A SB100x transposase vector mediated random integration of the tHMGA2/GFP transgene cassette. HSCs in mice were mobilized by subcutaneous injections of G-CSF and AMD3100/Plerixafor and intravenously injected with the integrating tHMGA2/GFP vector. This resulted in a slow but progressive, competitive expansion of GFP+ PBMCs, reaching about 50% by week 44 with further expansion in secondary recipients. Expansion occurred at the level of HSCs as well as at the levels of myeloid, lymphoid, and erythroid progenitors within the bone marrow and spleen. Importantly, based on genome-wide integration site analyses, expansion was polyclonal, without any signs of clonal dominance. Whole-exome sequencing did not show significant differences in the genomic instability indices between tHGMGA2/GFP mice and untreated control mice. Auto-expansion by tHMGA2 was validated in humanized mice. This is the first demonstration that simple injections of mobilization drugs and HDAd vectors can trigger auto-expansion of in vivo transduced HSCs resulting in transgene-marking rates that, theoretically, are curative for hemoglobinopathies.
Keywords: HMGA2; Sleeping Beauty transposase; gene therapy; helper-dependent adenovirus vector; hematopoietic stem cells; insertion site analysis; in vivo.
© 2024 The Author(s).
Conflict of interest statement
A.L. and P.N.V. receive research funding from Ensoma, Inc. R.E., K.W.M., and R.P. are employees of Ensoma, Inc.
Figures

Similar articles
-
In vivo HSC transduction in rhesus macaques with an HDAd5/3+ vector targeting desmoglein 2 and transiently overexpressing cxcr4.Blood Adv. 2022 Aug 9;6(15):4360-4372. doi: 10.1182/bloodadvances.2022007975. Blood Adv. 2022. PMID: 35679480 Free PMC article.
-
Targeted Integration and High-Level Transgene Expression in AAVS1 Transgenic Mice after In Vivo HSC Transduction with HDAd5/35++ Vectors.Mol Ther. 2019 Dec 4;27(12):2195-2212. doi: 10.1016/j.ymthe.2019.08.006. Epub 2019 Aug 19. Mol Ther. 2019. PMID: 31494053 Free PMC article.
-
HDAd6/35++ - A new helper-dependent adenovirus vector platform for in vivo transduction of hematopoietic stem cells.Mol Ther Methods Clin Dev. 2023 Mar 21;29:213-226. doi: 10.1016/j.omtm.2023.03.008. eCollection 2023 Jun 8. Mol Ther Methods Clin Dev. 2023. PMID: 37081854 Free PMC article.
-
Adenovirus vectors in hematopoietic stem cell genome editing.FEBS Lett. 2019 Dec;593(24):3623-3648. doi: 10.1002/1873-3468.13668. Epub 2019 Nov 20. FEBS Lett. 2019. PMID: 31705806 Free PMC article. Review.
-
Plerixafor: A chemokine receptor-4 antagonist for mobilization of hematopoietic stem cells for transplantation after high-dose chemotherapy for non-Hodgkin's lymphoma or multiple myeloma.Clin Ther. 2010 May;32(5):821-43. doi: 10.1016/j.clinthera.2010.05.007. Clin Ther. 2010. PMID: 20685493 Review.
References
-
- Yao J., Atasheva S., Wagner N., Di Paolo N.C., Stewart P.L., Shayakhmetov D.M. Targeted, safe, and efficient gene delivery to human hematopoietic stem and progenitor cells in vivo using the engineered AVID adenovirus vector platform. Mol. Ther. 2024;32:103–123. doi: 10.1016/j.ymthe.2023.10.023. - DOI - PMC - PubMed
-
- Ni S., Gaggar A., Di Paolo N., Li Z.Y., Liu Y., Strauss R., Sova P., Morihara J., Feng Q., Kiviat N., et al. Evaluation of adenovirus vectors containing serotype 35 fibers for tumor targeting. Cancer Gene Ther. 2006;13:1072–1081. - PubMed
-
- Richter M., Saydaminova K., Yumul R., Krishnan R., Liu J., Nagy E.E., Singh M., Izsvák Z., Cattaneo R., Uckert W., et al. In vivo transduction of primitive mobilized hematopoietic stem cells after intravenous injection of integrating adenovirus vectors. Blood. 2016;128:2206–2217. doi: 10.1182/blood-2016-04-711580. - DOI - PMC - PubMed
-
- Li C., Mishra A.S., Gil S., Wang M., Georgakopoulou A., Papayannopoulou T., Hawkins R.D., Lieber A. Targeted Integration and High-Level Transgene Expression in AAVS1 Transgenic Mice after In Vivo HSC Transduction with HDAd5/35++ Vectors. Mol. Ther. 2019;27:2195–2212. doi: 10.1016/j.ymthe.2019.08.006. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

