Single-Molecule Sequencing of the C9orf72 Repeat Expansion in Patient iPSCs
- PMID: 39282230
- PMCID: PMC11393041
- DOI: 10.21769/BioProtoc.5060
Single-Molecule Sequencing of the C9orf72 Repeat Expansion in Patient iPSCs
Abstract
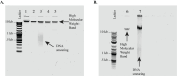
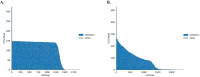
A hexanucleotide GGGGCC repeat expansion in the C9orf72 gene is the most frequent genetic cause of amyotrophic lateral sclerosis (ALS) and frontal temporal dementia (FTD). C9orf72 repeat expansions are currently identified with long-range PCR or Southern blot for clinical and research purposes, but these methods lack accuracy and sensitivity. The GC-rich and repetitive content of the region cannot be amplified by PCR, which leads traditional sequencing approaches to fail. We turned instead to PacBio single-molecule sequencing to detect and size the C9orf72 repeat expansion without amplification. We isolated high molecular weight genomic DNA from patient-derived iPSCs of varying repeat lengths and then excised the region containing the C9orf72 repeat expansion from naked DNA with a CRISPR/Cas9 system. We added adapters to the cut ends, capturing the target region for sequencing on PacBio's Sequel, Sequel II, or Sequel IIe. This approach enriches the C9orf72 repeat region without amplification and allows the repeat expansion to be consistently and accurately sized, even for repeats in the thousands. Key features • This protocol is adapted from PacBio's previous "no-amp targeted sequencing utilizing the CRISPR-Cas9 system." • Optimized for sizing C9orf72 repeat expansions in patient-derived iPSCs and applicable to DNA from any cell type, blood, or tissue. • Requires high molecular weight naked DNA. • Compatible with Sequel I and II but not Revio.
Keywords: C9orf72; No amplification; Repeat expansion; Single-molecule sequencing; iPSCs.
©Copyright : © 2024 The Authors; This is an open access article under the CC BY license.
Conflict of interest statement
Competing interestsJ.H. and Y-C.T. are full-time employees at Pacific Biosciences (PacBio) and J.H. holds stock in PacBio. C.D.C. is a founder, with equity, in Ciznor Co., a CNS therapeutics company. The other authors have no conflicts.
Figures
References
-
- DeJesus-Hernandez M., Mackenzie I. R., Boeve B. F., Boxer A. L., Baker M., Rutherford N. J., Nicholson A. M., Finch N. A., Flynn H., Adamson J., et al.(2011). Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9orf72 Causes Chromosome 9p-Linked FTD and ALS. Neuron. 72(2): 245-256. - PMC - PubMed
-
- Majounie E., Renton A. E., Mok K., Dopper E. G., Waite A., Rollinson S., Chiò A., Restagno G., Nicolaou N., Simon-Sanchez J., et al.(2012). Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 11(4): 323-330. - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

